Lecture 14 & 15
advertisement

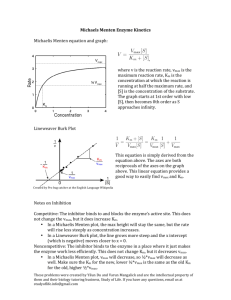
Lecture 14&15 Date: May 3&5 Chemistry 24b Spring Quarter 2004 Instructor: Richard Roberts Michaelis-Menten Kinetics k k k2 1 3 → E +P → → EP ← ES ← E+S ← k k k −2 −1 −3 Turnover is controlled by k2. Satisfied by initial rates or velocities and intial [ S] >> E0 , whence [ P] → 0 . Rate = velocity = v ≅ k2 [ES ] = where and k2 E0 [ S] Vmax [S ] = KM + [S ] K M + [S ] k−1 + k2 ⇐ Michaelis constant k1 = k2 E0 k2 is sometimes called kcat KM = Vmax hyperbolic kinetics Vmax v 1V 2 max KM [S] Figure 7-1 Kinetic Plots v = where V max [S] K M + [S] hyperbolic v ≡ initial rates [S] ≡ initial substrate concentrations >> E 0 Chemistry 24b — Lecture 14&15 2 Linearized Plots (1) Lineweaver-Burk: 1/v vs 1/[S] — Double Reciprocal 1 1 1 KM = + ⋅ V max v V max [S] 1 v Slope = KM /Vmax 1/Vmax 1/[S] −1 KM Figure 7-2 (2) Eadie-Hofster: v/[S] vs v — Single Reciprocal v V −v = max [S] KM Vmax /KM v [S] Slope = −KM−1 (for normal KM's) 0 Vmax Figure 7-3 v Chemistry 24b — Lecture 14&15 3 (3) Dixon: [S]/v vs [S] [ S] [S ] KM = + v V max V max [S] v Slope = 1 Vmax KM Vmax [S] 0 Figure 7-4 Inhibition of Enzymatic Reactions (1) Competitive Inhibition A competitive inhibitor is a molecule that resembles the substrate and occupies the catalytic site because of its similarity in structure, but is completely unreactive. By occupying the active site, the inhibitor prevents normal substrates from binding and being catalyzed. Operationally, competitive inhibitors bind reversibly to the active site. Hence, inhibition can be reversed by (1) diluting the inhibitor, or (2) swamping the system with excess substrate. Reaction Mechanism → → k1 k2 → ES ← → E + P E +S ← k−1 k−2 + I EI Define KI = [ E ][ I ] [EI ] Chemistry 24b — Lecture 14&15 4 Expectation A competitive inhibitor increases KM , but does not affect V max (because of sufficiently high [S], S will displace I). Hyperbolic Plots Vmax [I] = 0 v inhibitor KM' KM [S] Figure 7-5 v = V max [S] = [I] [S] + KM 1+ K V max [S] [S] + KM ′ I [I] where KM ′ = KM 1 + K I ⇐ modified K M Lineweaver-Burk Plots inhibitor 1 v [I] = 0 1/Vmax −1 KM −1 KM' Figure 7-6 1/[S] Chemistry 24b — Lecture 14&15 5 Eadie-Hofster Plot Vmax /KM v [S] Slope = −KM−1 inhibitor Vmax 0 v Figure 7-7 Dixon Plot parallel [S] v inhibitor KM Vmax [I] = 0 Slope = 1 Vmax [S] 0 Figure 7-8 Chemistry 24b — Lecture 14&15 6 Classical Example of Competitive Inhibition Enzyme: succinic dehydrogenase (SDH), which catalyzes the oxidation of succinic acid to fumaric acid. COOH CH2 COOH SDH CH Here CH 2 HC COOH substrate ≡ succinic acid inhibitor ≡ malonic acid COOH succinic acid COOH CH2 fumaric acid COOH Here KI = 1 × 10 −5 M , which means that at [malmate] = 10 −5 M , the apparent affinity of the enzyme for succinate decreases by a factor of 2! (2) Noncompetitive Inhibition A noncompetitive inhibitor is one that binds reversibly to the enzyme, but not at the active site itself, so that the substrate can still bind at the active site, but there’s no catalyzed transformation. This type of inhibition cannot be overcome by a large amount of substrate, thus noncompetitive inhibition. Reaction Mechanism k1 k2 → ES ← → E + P E +S ← k−1 k−2 + + I I → → → → . EI ES ⋅ I Define [E ][ I ] KI = [ EI ] eq [ES ][ I ] KSI = [ES ⋅ I ] eq Chemistry 24b — Lecture 14&15 7 Expectation The rate or velocity decreases to the extent that E is complexed by inhibitor, irrespective of EI or ES ⋅ I . Thus, a noncompetitive inhibitor decreases V max without affecting the apparent KM . The situation is not overcome by swamping the system with substrate. Hyperbolic Plots Vmax v no inhibitor Vmax ' inhibitor 1V max 2 1V ma ' x 2 [S] KM Figure 7-9 Typical Case KI = KSI , i.e., the affinity of the inhibitor site for the inhibitor does not depend on whether E is bound with S. In this instance v = where Vmax ′ = V max [S] = 1 + [I] ([S] + K ) M KI 1 1 + [I] KI ⋅ V max Vmax ′ [S] KM + [S] Chemistry 24b — Lecture 14&15 8 Lineweaver-Burk Plots 1 v inhibitor 1 Vmax ' no inhibitor 1 Vmax 1/[S] −KM−1 Figure 7-10 Eadie-Hofster Plot v [S] no inhibitor [I] lines parallel v 0 Figure 7-11 Chemistry 24b — Lecture 14&15 9 Dixon Plot (both slope and intercept increase) [I] 1 Vmax ' [S] v KM Vmax ' no inhibitor Slope = KM Vmax 1 Vmax [S] Figure 7-12 More General Case KI ≠ KSI allosteric interaction between catalytic and inhibitor sites. v = where V ′max [S] k2 E 0 [S] 1 ⋅ = α α [I] KM + [S] K + [S] 1+ β β M KSI [I] 1+ α KI = β 1 + [I] KSI and Both V max and KM can be affected! V ′max = Vmax β Chemistry 24b — Lecture 14&15 Case of 10 KSI >> KI This is an interesting situation where the allosteric interaction between catalytic and inhibitor sites is so strong that binding is mutually exclusive. v = If then V max [S] β α K + [S] β M 1 K SI >> [I] , v = β → 1 V max [S] = [I] 1+ K + [S] KI M V max [S] KM ′ + [S] which is the result for competitive inhibition! Irreversible Modification Permanent or irreversible modification of the active site often yields a situation that behaves like the case of simple noncompetitive inhibition! For example, irreversible modification by the chemical alkylating agent iodoacetamide, which reacts with exposed sulfhydryl groups such as a cysteine to form a covalently modified Enzyme — S — CH2 — C — NH 2 O or irreversible modification by carbodiimide of carboxyl groups from glutamate and aspartate. Chemistry 24b — Lecture 14&15 11 R O O NH R' Enzyme — C — O — C Enzyme — C — N N C R' NH O R Oftentimes, chemically modified enzyme is inactive catalytically. So, v ∝ [Enzyme ]active , i.e., enzyme not chemically modified and unreacted enzyme behave normally with KM identical to the situation prior to the addition of “inhibitor.” In fact v = f unmodified V max [S] K M + [S] like in simple noncompetitive inhibition. Well-Studied Enzyme System That Behaves According to Michaelis-Menten Kinetics Ferriprotoporphyrin and (Mn)2 protein catalase; catalyze the decomposition of H2O2. 2 H2 O2 → 2 H2 O + O2 Table 7-1. Velocities and Energies for Protein Catalases Catalyst None HBr Fe2+/Fe3+ Hematin or Hb Fe(OH)2 TETA+ b Catalase Velocitya, −d [H 2 O2 ] dt 10–8 10–4 10–3 10–1 103 107 ,M S −1 Ea (kJ/mole) 71 50 42 — 29 8 Chemistry 24b — Lecture 14&15 a b for [H 2O 2 ] = 1 M , triethylenetetramine 12 [catalyst] active sites = 1 M uncatalyzed Ε 71 kJ 8 kJ H2O2 −94.7 kJ H2O + 1/2 O2 Figure 7-13 0 ∆ G298 = − 103.10 kJ mol −1 for H2 O2 (aq) → H2 O (l) + 0 ∆ H298 = − 94.64 kJ mol 1 O2 (g) 2 −1 Turnover number (S −1 ) ≡ maximum velocity divided by the concentration of enzyme active sites Chemistry 24b — Lecture 14&15 13 Table 7-2. Turnover Numbers for Various Enzymes and Substrates Enzyme Substrate Catalase Acetylcholinesterase Lactate dehydrogenase (chicken) Chymotrypsin Myosin Fumarase H2O2 Acetylcholine Pyruvate Acetyl-L-tyrosine ethyl ester ATP L-Malate Fumarate CO2 HCO3− Carbonic anhydrase (bovine) *Typically, turnover number ~ 103 S−1 within a factor of 10. Turnover No. (S−1)* 9 × 106 1.2 × 104 6 × 103 4.3 × 102 3 1.1 × 103 2.5 × 103 8 × 104 3 × 104 Chemistry 24b — Lecture 14&15 14 Two Intermediate Complexes The reaction catalyzed by catalase is essentially irreversible. Therefore, it is not necessary to worry about EP. Most enzymatic reactions are readily reversible, so an enzyme-product complex can often be detected. Good Example COO H COO C − fumarase C + H2O OOC H—C—H HO — C — H C − − H COO − fumarate L-malate 20% 80% Equilibrium at room temperature k k2 k3 1 → ES ← → E P← →E + P E + S ← k k −1 k −3 −2 So in this case, you cannot ignore EP! More complete treatment leads to the following results: (1) Forward Reaction: [P]0 = 0 d[P] vF = = k3 [EP] dt 0 k2 k3 E0 [S] = k−1k −2 + k−1k3 + k 2k3 +[S] k1(k2 + k−2 + k3 ) (k2 + k−2 + k 3 ) which can be reduced to with vF = V F [S] F + [S] KM VF = k2 k3 E0 k2 + k−2 + k3 and F KM = k−1 k−2 + k −1k3 + k2 k3 k1 (k 2 + k−2 + k3 ) Chemistry 24b — Lecture 14&15 15 (2) Reverse Reaction: [S]0 = 01 d [S] vR = = k−1 [ES] dt 0 V R [P] K MR + [P] k−1 k−2 E0 VR = k−1 + k2 + k−2 = with and K MR = k−1 k−2 + k −1k3 + k 2 k3 k −3 (k −1 + k 2 + k−2 ) (3) Net Velocity R F [S] − V R KM [P] VF K M v = F R R F [P] K M K M + KM [S] + K M Note that at equilibrium, v = 0 and hence R F [S]eq = V R K M [P]eq V F KM and [P] V KR = F FM K = [S] eq V R KM There are too many kinetic constants here to be sorted out by steady state kinetics. We must appeal to the methods of rapid kinetics. 1Alberty and Peirce, J. Am. Chem. Soc. 79, 1526 (1957).