study session 4 Answers
advertisement

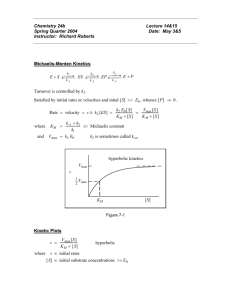
SCHOOL OF BIOLOGICAL SCIENCES Study Session 4 ENZYMOLOGY You should read chapter 8 (pps 189 - 225) of Biochemistry 5th Ed. Berg, Tymoczko and Stryer) and refer to your BS1090/BS1080 notes on enzymology before answering these questions. Questions 1(a) If AB is a strongly exergonic reaction why should A exist at all under normal conditions? Illustrate your answers with a diagram (energy changes during course of reaction). In the diagram show how an enzyme could increase the rate of formation of B. Transition State With enzyme G o Ea A Go B A Course of Reaction Ea lowered by enzyme, therefore formation of transition state facilitated. 1(b) How does your diagram illustrate that the equilibrium constant for the reaction is not changed? Go = -RTlnKeq Enzyme does not change overall Go, when A B Keq is not changed 1 (c) What is the active site of an enzyme? The 3-dimensional region of the protein to which substrates (and co-enzymes) bind to specific groups on the aminoacids at the active site. The site contains the catalytic groups of the protein which bring about the covalent changes during the reaction. (d) How do you explain the specificity of enzyme-catalysed reactions? Two models: (i) Lock-and –key (ii) Induced –fit substrate induces conformational change in enzyme e.g. hexokinase (e) The catalytic activity of an enzyme is pH-dependent. Draw a typical pH optimum curve for an enzyme-catalysed reaction and explain its shape. Maximal rate at optimum pH Vo pH At the optimum pH ionisable groups at the active site of the protein are in the correct state fror maximal catalytic activity 2(a) In what units are the rates of enzyme-catalysed reactions normally expressed? (i) Moles of substrate transformed (or product formed) per unit time e.g. mol. sec-1 (most commonly used in Biochemistry) SI unit = Katal = 1 mol.sec-1 (ii) 2 (b) Define the term, initial rate. How in practice is an initial rate normally measured? The rate at T0 . Usually measured by estimating the rate of reaction over time and then plotting data to ascertain the linear initial rate . (c) What is the specific activity of an enzyme? Why is it important in enzyme purifications? It is the number of enzyme units per fixed amount of protein. Moles of substrate transformed (or product formed) per unit time per mg protein e.g. mol. sec-1. mg(protein)-1 (most commonly used in Biochemistry). It is useful in calculating the purification of an enzyme (the higher the Specific Activity (SA), the greater the purification until a point is reached where the enzyme under consideration is the only protein left – in which case the SA will not change on subsequent attempts at purification) Do not confuse with Total Activity (mol. sec-1) or % Recovery 3. Draw a graph to illustrate change in V0 with increasing [S], keeping [E] constant. Indicate Vmax on the graph. Vmax First Order Kinetics V0 Zero Order Kinetics [S] E + S E-S E + P At Vmax , all the E is present as the E-S complex 3 4. The Michaelis-Menten equation is Vmax [S] V0 = ----------[S] + Km ____________ (1) Define Km (the Michaelis constant) in terms of the rate constants for the formation and breakdown of the ES complex. What is the practical value of calculating the Km? k1 k3 E + S ES E + P k2 k 2 k3 Km ( note that Km has units of concentration, M, mM, M etc.) k1 Km is equal to the substrate concentration [S] at half of the maximum velocity. It reflects the stability of the E-S complex (or how tightly the enzyme binds substrate. Therefore High Km = low affinity of E for S; Low Km indicates high affinity) 5. Lineweaver-Burk Equation (Double reciprocal plot) Vmax (maximum rate when E is fully saturated with S) and Km values for an enzyme can be calculated using the Lineweaver-Burk equation (2): 1 1 Km 1 -- = -- + ---- . --V0 Vmax Vmax [S] ____________ (2) Draw a typical Lineweaver-Burk plot indicating all the terms in equation (2). How is Km calculated from the plot? I/V0 Slope = Km/Vmax I/Vmax 6. 1/[S] Define the term “turnover number”, Kcat . What information does it provide? -1/Km 4 Kcat is the maximum number of substrate molecules that can be converted to product per second and per active site. It is ameasure of the efficiency of the enzyme – how rapidly an enzyme can catalyse a reaction once the substrate has bound to the active site. Some enzymes are much more efficient than others (e.g. Catalase 4 x 107mol/sec; lysozyme 0.5 mol/sec). 7. Competitive and non-competitive inhibition can be recognised by Lineweaver-Burk plots (equation (2)) of the enzyme-catalysed reaction in the absence and presence of the inhibitor. (a) Plot 1 1 --- against ---- for an uninhibited reaction and competitively inhibited reaction V [S] at two different concentrations of inhibitor , and explain what happens to Km and Vmax values. Vmax is not affected by competitive inhibition. Km is altered (increased) because I and S compete for the same (active) site on the enzyme. +I2 1/V0 +I1 Control 1/Vmax 1/[S] -1/Km 5 7(b) What sorts of compounds commonly inhibit enzyme-catalysed reactions competitively and why? Compounds of structural similarity to the substrate that can therefore bind at the same site on the enzyme e.g. malonate inhibits succinate dehydrogenase Malonate CO2H. CH2. CO2H Succinate CO2H. CH2. CH2. CO2H (c) Draw a Lineweaver-Burk plot to indicate the effect of a non-competitive inhibitor (2 concentrations) on an enzyme-catalyzed reaction, and show Km and Vmax in each case. Increasing [I] +I2 +I1 1/v0 Control 1/vmax changes 1/[S] -1/km constant Vmax decreased as [I} increases Km is unaffected 6 (d) Give an example of a non-competitive inhibitor. How does its mode of action differ from a competitive inhibitor? galactosidase inhibited by Ag2+ ions Fructose bisphosphatase inhibited by AMP Inhibitor is not structurally related to the substrate Does not bind to the same site as substrate but interferes with the formation of the E-S complex Causes some conformational change in the protein No change in Km of enzyme (e) Competitive and non-competitive inhibition are reversible. Name an irreversible inhibitor and explain its reaction with the enzyme. Usually forms a covalent bond with an aminoacid side chain or with a bound coenzyme. e.g. cyanide, p-mercuribenzoate, iodoacetate these can change Km, Vmax or both. DRD/study session 4/ /3.02.04 7