Outline
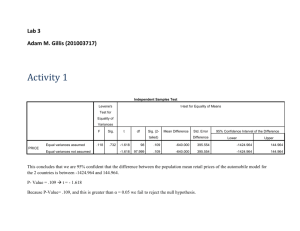
advertisement

Outline
1
Descriptive Statistics: Box plots
2
Comparing Two Samples
Paired samples
Independent samples - equal variances
Independent samples - unequal variances
3
Nonparametric methods for two samples
Levene’s test
Mann-Whitney test
4
Comparing two proportions
With a z-test
Confidence interval
With a chis-square test
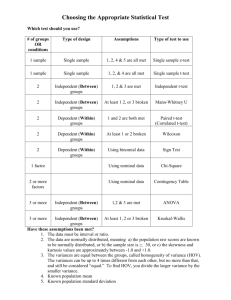
1
The sample median
To compute the sample median:
arrange the data in a list of ascending order
take the middle value in the list
(
ỹ
=
n+1 th
2
avg of
value
n th
2
and
n
2
th
+1
value
if n is odd
if n is even
Height of red pine seedlings from the nursery: ỹ =
2
3
4
5
6
|
|
|
|
|
cm.
36
144
23679
46
1
If n is even, the sample median is the average of the
middle two values.
Ex: median of 2,4,5,7,8,14 is:
2
Sample quantiles
The median is a special quantile (or percentile): ỹ = y[.50]
To compute the pth sample quantile y[p] (0 ≤ p ≤ 1):
arrange data in a list of ascending order
compute np
If np is an integer, then y[p] is the average of (np)th and
(np + 1)th data values in the list.
If np is not an integer, then round up to [np] and use the
[np]th data value in the list.
Ex: height of seedlings, n = 13. The 0.20th sample
quantile, or 20th percentile is y[0.20] =
p = .1538: y[0.1538] =
(note: np = 2)
3
Box plots
y[0.50] = sample median
y[0.25] = first quartile (Q1)
y[0.75] = third quartile (Q3)
Height of seedlings: y[0.25] =
y[0.75] =
and
W&S and other texts provide a different definition:
Q1 as the median of the first half (median excluded),
Q3 as the median of the second half of the data.
They are not equivalent definitions (check?).
A box plot displays several quantiles simultaneously:
4
Sample range and IQR
Sample range
Sample range = largest obs − smallest obs,
is a measure of spread/variability of the data set.
Height of seedlings: sample range is:
Interquartile range (IQR)
Interquartile range = difference between the third quartile
and the first quartile: IQR= Q3 − Q1 = y[0.75] − y[0.25]
For the height of seedlings, the IQR is:
5
Box plot for production of organic cows
Milk yield data
IQR: 14
Q1
Min
Median
Q3
Max
Range: 36
20
25
30
35
40
45
50
55
6
Box plots are useful to compare samples
Fungus colonization from 3 samples: black mustards producing
a large/low amount of sinigrin, and other plant species.
●
% fungus colonization
12
10
8
6
4
2
heterospecific high sinigrin
low sinigrin
7
R commands
#
>
>
>
+
+
enter data:
hi = c(4.43,2.18,6.64,4.41,3.7,4.79,3.38,8.37,2.94,6.92,7.24)
lo = c(11.07,7.89,7.89,8.12,8.11,10.79,10.3,7.21,5.77,10.47,8.09)
het = c(7,7.03,9.17,6.75,3.45,6.63,4.52,8.09,7.55,6.63,8.68,9.4,
5.22,9,3.96,5.46,7.75,9.11,9.01,7.46,6.41,5.29,3.65,8.29,9.55,
4.3,6.45,6.46,4.83,7.98,5.64,6.85,13.41)
> summary(hi)
Min. 1st Qu.
2.18
3.54
> IQR(hi)
[1] 3.24
Median
4.43
# summary stats, high sinigrin sample
Mean 3rd Qu.
Max.
5.00
6.78
8.37
# IQR
> boxplot(hi)
> boxplot(hi,lo,het)
# boxplot, high sinigrin only
# boxplot, all 3 samples side-by-side
# same but group names added:
> boxplot(hi,lo,het, names=c("High sinigrin","Low","Heterospecific"))
#
>
>
>
if data is read from a file:
dat = read.csv("blackmustard.csv", header=TRUE)
dat
boxplot(fungus_percent ˜ community, data=dat)
8
Outline
1
Descriptive Statistics: Box plots
2
Comparing Two Samples
Paired samples
Independent samples - equal variances
Independent samples - unequal variances
3
Nonparametric methods for two samples
Levene’s test
Mann-Whitney test
4
Comparing two proportions
With a z-test
Confidence interval
With a chis-square test
9
Paired vs. Independent samples
Treatments: A and B.
Paired samples: each observation on trt A is naturally
paired with an observation on trt B. Related or same
experimental units are used for both treatments.
Independent samples: no direct relationship between an
observation on trt A and an observation on trt B.
Choice of paired versus independent sample is an important
design issue. Data analysis follows the design.
10
Examples
Two-sample comparisons are very common. Examples:
1
Compare fungus colonization in low/high sinigrin mustard
2
Compare taste of cheese from cows on two different diets
(organic in the open vs. non-organic, hay/pellets)
3
Compare cholesterol level of patients before and after a
drug treatment
4
Baby weight at birth among smoking/non-smoking women
11
When, why should samples be paired?
Cholesterol example:
1
Cholesterol level of 10 patients before and after a drug
treatment.
2
Cholesterol level of 10 patients before treatment and of
another 10 patients after treatment.
Baby weight example: pairing women according to certain
traits. Effective only if it controls variability.
Paired sample studies usually preferred, because of
increased precision (i.e. reduced variability) in estimating
treatment differences.
If 3 or more treatments, blocking replaces pairing.
12
Paired samples - Blood pressure example
Question of interest: is there any evidence that a particular
drug has an effect on blood pressure?
Experiment: on 15 middle-aged male hypertension patients.
For each patient, blood pressure is measured at time of
enrollment and again after 6 months of the drug treatment.
13
Blood pressure (mm Hg)
Subject
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
Before (Y1 )
90
100
92
96
96
96
92
98
102
94
94
102
94
88
104
After (Y2 )
88
92
82
90
78
86
88
72
84
102
94
70
94
92
94
Difference (D = Y1 − Y2 )
2
8
10
6
18
10
4
26
18
-8
0
32
0
-4
10
14
Blood pressure example
µ1 = population mean blood pressure before the drug trt
µ2 = population mean blood pressure after the drug trt
µD = µ1 − µ2 = difference between the two mean blood
pressure levels.
Paired samples
Testing µ1 = µ2 or µ1 6= µ2 is equivalent to testing
H0 : µD = 0 vs HA : µD 6= 0.
A one-sample t-test can be used on the differences.
The difference D = Y1 − Y2 is the blood pressure difference.
We assume we have a random sample D1 , D2 , . . . , D15 of size
n = 15 from N (µD , σD2 ).
15
Blood pressure example
Under H0 : µD = 0, the test statistic T =
D̄ − 0
√ has a
SD / n
t-distribution on df =
We observed d̄ = 8.80 mm Hg, sd = 10.98. The observed
t-value is
= 3.10
t=
on df =
The p-value is 2IP{T ≥ 3.10}, which is between 0.002 and
0.01 from Table C.
There is strong evidence against H0 : the drug is deemed ...
16
Confidence interval from paired samples
A (1 − α) CI for the difference of means µ1 − µ2 = µD is
s
s
d̄ − tn−1,α/2 √d ≤ µD ≤ d̄ + tn−1,α/2 √d
n
n
Blood pressure:
≤ µD ≤
We are 95% confident that the population mean decrease in
blood pressure after 6 months of treatment lies between 2.72
and 14.88 mm Hg (or 8.80 ± 6.08).
Remarks
If a difference of 5 mm Hg is needed for biological
significance, we could test H0 : µD = 5 vs. HA : µD 6= 5.
Assumptions: random sample, and D values have normal
distribution. Check with a normal probability plot
No normality assumption about Y1 , or about Y2 . Usually
not independent due to pairing.
17
R commands
>
>
>
>
>
>
# first enter the data
bpbefore =c(90,100,92,96,96,96,...,102,94,88,104)
bpafter = c(88, 92,82,90,78,86,..., 70,94,92, 94)
# Now do the paired t-test and 95% CI
t.test( bpbefore - bpafter )
One Sample t-test
data: bpbefore - bpafter
t = 3.1054, df = 14, p-value = 0.00775
alternative hypothesis: true mean is not equal to 0
95 percent confidence interval:
2.722083 14.877917
sample estimates:
mean of x
8.8
18
R commands
> # or better use of the function t.test:
>
> t.test(bpbefore, bpafter, paired=TRUE)
Paired t-test
data: bpbefore and bpafter
t = 3.1054, df = 14, p-value = 0.00775
alternative hypothesis: true difference in means is not
95 percent confidence interval:
equal to 0
2.722083 14.877917
sample estimates:
mean of the differences
8.8
19
Independent samples
Compare mycorrhizal colonization in soil from high-sinigrin
black mustard communities (11
rep) and low-sinigrin black mustard communities (11 rep).
Question of interest: is there
evidence of an effect of black
mustard (high/low sinigrin) on
fungus colonization?
20
Mycorrhizal colonization example
Data: mycorrhizal colonization (% of root section), in
high-sinigrin (hi) and low-sinigrin (lo) communities.
community
1
2
3
4
5
6
7
8
9
10
11
hi (Y1 )
4.43
2.18
6.64
4.41
3.70
4.79
3.38
8.37
2.94
6.92
7.24
lo (Y2 )
11.07
7.89
7.89
8.12
8.11
10.79
10.30
7.21
5.77
10.47
8.09
No pairing: observations can
be permuted within each trt
(column).
high
sinigrin
low
sinigrin
2
3
4
5
6
7
8
9
% fungus colonization
10
11
ȳ1 = 5, s1 = 2.0
ȳ2 = 8.7, s2 = 1.7
21
Mycorrhizal colonization
Let µ1 = the population mean fungus colonization in
communities assigned to high-sinigrin black mustard,
µ2 = the population mean fungus colonization with
low-sinigrin.
We want to test H0 : µ1 = µ2 versus HA : µ1 6= µ2 .
µ1 = µ2 means µ1 − µ2 = 0.
Main idea: use Ȳ1 − Ȳ2 , because IE Ȳ1 − Ȳ2 = µ1 − µ2 .
If Ȳ1 − Ȳ2 is close to 0, we will
if Ȳ1 − Ȳ2 is far from 0, we will
Here ȳ1 − ȳ2 = 5 − 8.7 = −3.7 (% root section difference).
22
Assumptions
1
2
3
Two independent random samples Y1 and Y2 .
The first sample Y11 , Y12 , . . . , Y1n1 is from N (µ1 , σ12 ),
second sample Y21 , Y22 , . . . , Y2n2 is from N (µ2 , σ22 ).
The variances are the same σ12 = σ22 = σ 2 .
Essentially, three assumptions: independence (within a trt and
between two trts), normality, and equal variance.
No need to have equal sample size.
23
Distribution of Ȳ1 − Ȳ2
Under these assumptions
Ȳ1 ∼ N
and Ȳ2 ∼ N
If H0 is true, Ȳ1 − Ȳ2 has a normal distribution with
mean: IE Ȳ1 − Ȳ2 = µ1 − µ2 = 0
variance:
var(Ȳ1 −Ȳ2 ) =
=σ
2
1
1
+
n1 n2
But... how do we know σ 2 ?
24
S12 estimates σ 2 and so does S22 . A pooled estimate of σ 2 is
Pooled estimated of σ 2
Sp2 =
(n1 − 1)S12 + (n2 − 1)S22
sum of all deviations2
=
n1 + n2 − 2
n1 + n2 − 2
weighted average of S12 and S22 , weighted by the df’s.
Now we can estimate σȲ1 −Ȳ2 by
Standard error of Ȳ1 − Ȳ2
s
SȲ1 −Ȳ2 = Sp
1
1
+
n1 n2
With equal sample sizes n1 = n2 = n we get
r
Sp2 = (S12 + S22 )/2 and SȲ1 −Ȳ2 = Sp
2
n
25
The two-sample t-test
1
2
Hypotheses: H0 : µ1 = µ2 versus HA : µ1 6= µ2
Test statistic:
T =
Ȳ1 − Ȳ2 − 0
SȲ1 −Ȳ2
If H0 is really true then T ∼ t-distribution with df= n1 + n2 − 2
3
Get the data and p-value: ȳ1 − ȳ2 = 5 − 8.7 % root section,
s1 = 2.0 and s2 = 1.7, with n1 = n2 = 11
Pooled estimate of σ:
Standard error of Ȳ1 − Ȳ2 :
t-value:
4
sp =
= 1.856
t=
= .791
= 4.67
, p-value: 2IP{T20 > 4.67} < .001
Finally, df=
We 2 accept 2 reject H0 . Or: There is
evidence that fungus colonization is affected by the type of
black mustard community.
26
Confidence intervals
Independent samples
A (1 − α) confidence interval for µ1 − µ2 is
s
1
1
ȳ1 − ȳ2 ± t ∗ sp
+
n1 n2
where the multiplier t = tn1 +n2 −2,α/2 is determined by the
t-distribution with n1 + n2 − 2 df.
fungus colonization: a 99% CI for µ1 − µ2 is (using t = 2.845)
which is −3.7 ± 2.25 or [−5.95, −1.45] % of root sections.
The test and the CI are consistent:
27
R commands
>
>
>
>
>
>
# enter the data:
lo = c(11.07,7.89,7.89,8.12,...,7.21,5.77,10.47,8.09)
hi = c( 4.43,2.18,6.64,4.41,...,8.37,2.94, 6.92,7.24)
# do the test:
t.test(hi, lo, var.equal=T, conf.level=.99)
Two Sample t-test
data: hi and lo
t = -4.6786, df = 20, p-value = 0.0001444
alternative hypothesis: true difference in means is not
99 percent confidence interval:
-5.951651 -1.450167
sample estimates:
mean of x mean of y
5.000000 8.700909
28
Another example
Compare fungus colonization in high-sinigrin and in
heterospecific communities (mixed species, no black mustard).
ȳhet = 7.0, shet = 2.1, nhet = 33
ȳhi = 5.0, shi = 2.0, nhi = 11
ȳlo = 8.7, slo = 1.7, nlo = 11.
●
% fungus colonization
12
10
Test µhi = µhet .
8
6
4
2
heterospecific high sinigrin
low sinigrin
29
T-test with unequal variances
Some soaps are labelled “antibacterial” soaps, but one might
expect ordinary soap also to kill bacteria. Experiment: prepare
solution from regular soap and solution of sterile water.
Solutions were placed onto petri dishes along with E. coli
bacteria, incubated for 24h.
Question: is regular soap solution preventing growth of
bacteria compared to sterile water?
Control
30
36
66
21
63
38
35
45
Soap
76
27
16
30
26
46
6
Data: # bacteria colonies on each dish
n
mean
sd
Control
8
41.75
15.6
Soap
7
32.43
22.8
No pairing.
We refuse to assume σ1 6= σ2 .
30
ȳ1 − ȳ2 = 9.32, but how big could that be by chance alone?
Test of H0 : µ1 = µ2 with unequal variances
s
S12 S22
Ȳ1 − Ȳ2
with SȲ1 −Ȳ2 =
+
.
n1
n2
SȲ1 −Ȳ2
The p-value is obtained by comparing the value of T with a
t-distribution with adjusted degree of freedom
Test statistic: T =
df =
(v1 + v2 )2
v21
n1 −1
+
v22
n2 −1
where v1 = S12 /n1 and v2 = S22 /n2 .
df will always be at most n1 + n2 − 1 and always be ≥ the
minimum of n1 − 1 and n2 − 1.
df will not necessarily be an integer.
31
Soap experiment
ȳ1 − ȳ2 = 9.32 more colonies in water than in soap.
We get v1 =
sȳ1 −ȳ2 =
= 30.42, v2 =
= 74.26, then
= 10.25 colonies and df= 10.4.
t=
We use Table C with df=
a two-sided test.
= 0.909
and get p-value > 0.20 with
There is no evidence of antibiotic effect in soap (p> .2)
from this experiment.
By comparison: the t-test assuming equal variances yields
sȳ1 −ȳ2 = 9.98 colonies, t = 0.933, which would be
compared to the t-distribution on df=
. Same
conclusion.
32
Confidence interval for µ1 − µ2 with unequal variances
A (1 − α) confidence interval for µ1 − µ2 is
ȳ1 − ȳ2 ± t ∗ sȳ1 −ȳ2
where t = tdf,α/2 and df is the adjusted degree of freedom,
and
s
s12
s2
+ 2.
sȳ1 −ȳ2 =
n1 n2
In the soap example, for 90% confidence we get
t=
= 1.812 and interval:
i.e. [-9.2, 27.9] more colonies on average in water than in soap.
33
Which t-test should I use?
Assumptions (t-test not assuming equal variances)
1
Independence, within and among samples,
2
Each sample comes from a Normal distribution or is large
enough.
If software allows, use the t-test not assuming equal
variances by default.
If variances turn out to be significantly different (from
Levene’s test): find the right software!
On exams, use t-test assuming equal variances, and test
this assumption. Unless indicated otherwise.
34
Outline
1
Descriptive Statistics: Box plots
2
Comparing Two Samples
Paired samples
Independent samples - equal variances
Independent samples - unequal variances
3
Nonparametric methods for two samples
Levene’s test
Mann-Whitney test
4
Comparing two proportions
With a z-test
Confidence interval
With a chis-square test
35
Assessing assumptions
Assumptions of the independent two-sample t-test?
The t-test assuming equal variances is
robust against nonnormality, but sensitive to dependence.
moderately robust against unequal variance (σ12 6= σ22 ) if
n1 ≈ n2 , but much less robust if n1 and n2 are quite
different (e.g. differ by a ratio of 3 or more).
How to determine whether the equal variance assumption is
appropriate?
Chi-square test to compare σ12 and σ22 using S12 and S22 , but
avoid it: very sensitive to nonnormality.
Levene’s test: nonparametric test for comparing two
variances. Does not assume normality, still assumes
independence.
36
Levene’s test
Consider two independent samples Y1 and Y2 :
Sample 1: 4, 8, 10, 23
Sample 2: 1, 2, 4, 4, 7
Test H0 : σ12 = σ22 vs. HA : σ12 6= σ22 .
Sample variances: s12 = 67.58, s22 = 5.30.
Main idea of Levene’s test: turn testing for equal variances
using the original data into testing for equal means using
modified data.
Suppose normality and independence, if Levene’s test
gives a small p-value (< 0.01), then we use the
approximate test for H0 : µ1 = µ2 vs. HA : µ1 6= µ2 that
does not require equal variances.
37
Levene’s test
1
Find the median for each sample. Here ỹ1 = 9, ỹ2 = 4.
2
Subtract the median from each obs
Sample 1: -5, -1, 1, 14
Sample 2: -3, -2, 0, 0, 3
3
Take absolute values: we get deviations (not the usual
ones) with positive signs.
Sample 1*: 5, 1, 1, 14
Sample 2*: 3, 2, 0, 0, 3
4
For any sample with odd sample size, remove 1 zero.
Sample 1*: 5, 1, 1, 14
Sample 2*: 3, 2, 0, 3
5
Perform an independent two-sample t-test on the modified
samples.
38
Levene’s test
Here ȳ1∗ = 5.25, ȳ2∗ = 2, s12∗ = 37.58, s22∗ = 2.00.
We get sp2 =
= 19.79, sp = 4.45 and
t=
on df=
= 1.03
. The p-value 2 P(T6 ≥ 1.03) is > 0.20.
Do not reject H0 . Going back to the original samples, could we
use the t-test that assumes equal variances?
39
R commands
There is no predefined function to do Levene’s test in R, but we
can just copy and paste a function available from the course
website.
> y1 = c(4,8,10,23); y2 = c(1,2,4,4,7)
> levene.test(y1, y2)
Two Sample t-test
data: levene.trans(data1) and levene.trans(data2)
t = 1.0331, df = 6, p-value = 0.3414
alternative hypothesis: true difference in means is not
95 percent confidence interval:
-4.447408 10.947408
sample estimates:
mean of x mean of y
5.25
2.00
40
Mann-Whitney test (aka Wilcoxon rank sum test)
What if one small sample (or both) are not normally distributed?
Mann-Whitney: Non-parametric test for two independent
samples.
Analogous test exists for paired samples (signed rank test).
No distribution assumption, but still assume independence.
Main idea: look at the ranks of the observations
Consider two independent samples Y1 and Y2 :
Sample 1: 11, 22, 14, 21
Sample 2: 20, 9, 12, 10
Test H0 : µ1 = µ2 vs. HA : µ1 6= µ2 .
41
Mann-Whitney test
1
2
Rank the observations:
rank
obs
sample
1
2
3
4
5
6
7
8
9
10
11
12
14
20
21
22
2
2
1
2
1
2
1
1
Compute the sum of ranks
for each sample. Here
RS1 =
= 23
RS2 =
= 13
keep the smallest T = 13.
3
Under H0 the means are
equal, the rank sums should
be ∼ equal, so the smallest
should not be too small. How
small just by chance?
p-value=IP{T ≤ 13}=2IP{RS2 ≤ 13}
To compute it, we list all possible
orderings and get the rank sum of
each possibility.
IP{RS2 ≤ 13} =
so p-value = 0.2.
42
Mann-Whitney test
Rankings with RS2 ≤ 13:
rank
sample number
1
2
3
4
5
6
7
8
2
2
2
2
1
1
1
1
2
2
2
1
2
1
1
1
2
2
2
1
1
2
1
1
2
2
2
1
1
1
2
1
2
2
1
2
2
1
1
1
2
2
1
2
1
2
1
1
2
1
2
2
2
1
1
1
RS2
10
11
12
13
12
13
13
Total numbers of rankings:
43
Mann-Whitney test
Had we observed T = 10, p-value = 2 ∗ 1/70 = 0.0286.
Had we observed T = 11, p-value = 2 ∗ 2/70 = 0.0571.
With this sample size, we can only reject at 5% if the observed
rank sum is 10, i.e. all values in one sample are ...
A table (see course webpage) gives the cut-off values for
different sample sizes.
For n1 = n2 = 4 and α = 0.05, we can only reject H0 if the
observed rank sum is 10.
44
R command: wilcox.test
> samp1 = c(11, 22, 14, 21)
> samp2 = c(20, 9, 12, 10)
>
> wilcox.test(samp2, samp1)
Wilcoxon rank sum test
data: samp2 and samp1
W = 3, p-value = 0.2
alternative hypothesis: true mu is not equal to 0
45
Mann-Whitney test with unequal sample sizes
Recorded below are the longevity of two breeds of dogs.
Breed A
Breed B
obs
12.4
15.9
11.7
14.3
10.6
8.1
13.2
16.6
19.3
15.1
rank
obs
11.6
9.7
8.8
14.3
9.8
7.7
rank
n2 = 10
104.5
n1 = 6
T ∗ = 31.5
46
Mann-Whitney test
n1 = smaller sample size, n2 = larger sample size.
T ∗ = sum of ranks in the smaller group. Let
T ∗∗ = n1 (n1 + n2 + 1) − T ∗ = 6 × 17 − 31.5 = 70.5 and
T = min(T ∗ , T ∗∗ ) = 31.5
The p-value is 2IP{T ≤ 31.5|H0 } =?
Look up the table: t = 31.5 is between 27 and 32, the
p-value is between 0.01 and 0.05. Reject H0 at 5%.
Remarks
If there are ties, the table gives approximation only.
The test does not work well if the variances are very
different.
47
R command: wilcox.test
> breedA = c(12.4, 15.9, 11.7, 14.3, 10.6, 8.1, 13.2,
+
16.6, 19.3, 15.1)
> breedB = c(11.6, 9.7, 8.8, 14.3, 9.8, 7.7)
> wilcox.test(breedB, breedA)
Wilcoxon rank sum test with continuity correction
data: breedB and breedA
W = 10.5, p-value = 0.03917
alternative hypothesis: true mu is not equal to 0
Warning message:
Cannot compute exact p-value with ties in:
wilcox.test.default(breedB, breedA)
48
Outline
1
Descriptive Statistics: Box plots
2
Comparing Two Samples
Paired samples
Independent samples - equal variances
Independent samples - unequal variances
3
Nonparametric methods for two samples
Levene’s test
Mann-Whitney test
4
Comparing two proportions
With a z-test
Confidence interval
With a chis-square test
49
Comparing Two Proportions
Association genotype - phenotype: cross 2 inbred lines of mice,
one lean, one naturally obese. Backcross with the lean parent:
F2 mice.
genotype at a given locus: either LO or LL.
Phenotype: either lean or obese.
pLL = p1 probability of obese phenotype among F2
backcrosses with genotype LL at the locus,
pLO = p2 probability of obese phenotype among F2
backcrosses with genotype LO at the locus.
50
Test for comparing two proportions
We want to test H0 : p1 = p2 versus HA : p1 6= p2 and the data
will be Y1 ∼ B(n1 , p1 ) and Y2 ∼ B(n2 , p2 ) independent.
Use the difference in sample proportions
p̂1 − p̂2 =
Y1 Y2
−
.
n1
n2
IE(p̂1 − p̂2 ) = p1 − p2 and
var(p̂1 − p̂2 ) =
Under H0 : p1 = p2 = p, we have IE(p̂1 − p̂2 ) = 0 and
var(p̂1 − p̂2 ) = p(1 − p)(1/n1 + 1/n2 ), so that
Z =
≈ N (0, 1)
But p is still unknown. We estimate it by
51
Test for comparing two proportions
Test statistic
Y1 + Y2
p̂1 − p̂2 − 0
using p̂ =
Z =p
n1 + n2
p̂(1 − p̂)(1/n1 + 1/n2 )
Z is approximately N (0, 1) under H0 .
Data: nLO = 105, YLO = 71 mice with genotype LO are obese,
nLL = 87, YLL = 45 with genotype LL are obese.
52
Phenotype-genotype association
Here p̂LO =
= 0.676, p̂LL =
estimate of obesity rate is
= 0.517, the pooled
p̂ =
= 0.604
√
= 0.0056 = 0.075
SEpLO −pLL =
Observed test statistic is
z=
= 2.24
We compare z = 2.24 to N (0, 1): the p-value is
2 IP{Z ≥ 2.24} = 0.025.
Reject H0 at the 5% level, or
evidence against H0 .
53
Confidence interval
Recall var(p̂1 − p̂2 ) =
Here we don’t assume p1 = p2 , we just plug in p̂1 and p̂2 .
Confidence interval for p1 − p2 , with (1 − α) confidence
s
p̂1 − p̂2 ± zα/2
p̂1 (1 − p̂1 ) p̂2 (1 − p̂2 )
+
n1
n2
Obesity rate: 95% confidence interval for pLO − pLL :
i.e. 0.159 ± 0.138 or [0.021, 0.297].
We are 95% confident that genotype LO is associated with an
increase of obesity rate (compared to LL) by a value between
2.1 and 29.7 percentage points.
54
Comparing two proportions - sample size requirement
Need to check that the normal approximation works well:
For the confidence interval for p1 − p2 , check that
n1 p̂1 ≥ 5, n1 q̂1 ≥ 5, n2 p̂2 ≥ 5 and n2 q̂2 ≥ 5.
That’s easy...
In testing H0 : p1 = p2 , check that n1 p̂ ≥ 5, n1 q̂ ≥ 5,
n2 p̂ ≥ 5 and n2 q̂ ≥ 5.
55
R command: prop.test
> prop.test(c(71, 45), c(105, 87), correct=F)
2-sample test for equality of proportions without
continuity correction
data: c(71, 45) out of c(105, 87)
X-squared = 5.0264, df = 1, p-value = 0.02496
alternative hypothesis: two.sided
95 percent confidence interval:
0.02097751 0.29692068
sample estimates:
prop 1
prop 2
0.6761905 0.5172414
56
R command: prop.test
> prop.test(c(71, 45), c(105, 87))
2-sample test for equality of proportions with
continuity correction
data: c(71, 45) out of c(105, 87)
X-squared = 4.3837, df = 1, p-value = 0.03628
alternative hypothesis: two.sided
95 percent confidence interval:
0.01046848 0.30742971
sample estimates:
prop 1
prop 2
0.6761905 0.5172414
57
Chi-square test of independence/association
Are phenotype and genotype at a given locus independent?
associated?
LO
LL
total
71 45
116
lean
34
42
76
total
105 87
192
obese
He want to test H0 : pLO = pLL against HA : pLO 6= pLL .
Equivalently:
H0 : genotype and obesity phenotype are independent.
HA : genotype at the locus and phenotype are not independent:
one genotype tends to be associated with one phenotype.
58
Chi-square test of independence/association
1
Build table of expected counts under H0 .
If H0 is true, pLO = pLL , but we don’t know this value. Our best
guess:
p̂ =
total # successes
=
total # trials
somewhere between p̂LO =
= .60
= .68 and p̂LL =
= .52.
Expected # obese mice with LO genotype:
116
105 ∗ p̂ = 105 ∗ 192
= 63.44.
In general,
E=
Row total * Column total
Grand total
59
Chi-square test of independence/association
Observed counts:
LO
Expected counts when genotype
and phenotype are independent:
LO
LL
total
LL
total
71 45
116
obese
63.44 52.56
116
lean
34
42
76
lean
41.56 34.44
76
total
105 87
192
total
105
192
obese
87
Calculate the test statistic X 2 , distance to independence:
X (obs − exp)2
X2 =
=
exp
2
all cells
= 5.026
60
Test of independence
3
p-value: If there is independence (phenotype does not
depend on genotype) then X 2 has a χ2 distribution with
df= 1 here.
Using Table B: .02 < p < .05.
4
Conclusion: There is moderate evidence that the
genotypes have different obesity rates.
Furthermore, in the data we have p̂1 = .68 > p̂2 = .52.
There is moderate evidence that LO has a higher obesity
rate.
Same conclusion as a z test for testing pLL = pLO . We had
z = 2.25. Here X 2 = 5.026 = z 2 and exact same p-value.
61
Using R: chisq.test()
> mice = matrix( c(71,34,45,42), 2,2)
> mice
[,1] [,2]
[1,]
71
45
[2,]
34
42
> chisq.test(mice)
Pearson’s Chi-squared test with
Yates’ continuity correction
data: mice
X-squared = 4.3837, df = 1, p-value = 0.03628
> chisq.test(mice, correct=FALSE)
Pearson’s Chi-squared test
data: mice
X-squared = 5.0264, df = 1, p-value = 0.02496
62