IV. -Amino Acids: carboxyl and amino groups bonded to
advertisement

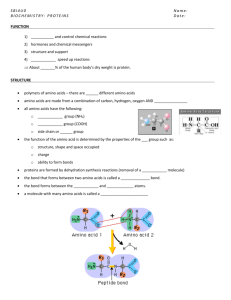
IV. -Amino Acids: carboxyl and amino groups bonded to -Carbo n A. Acid/Base properties 1. carboxyl group is proton donor ! weak acid 2. amino group is proton acceptor ! weak base 3. At physiological pH: H3N+-C -C O O B. Ca is tetrahedral and bonded to 4 different groups 1. L configuration for all natural amino acids (few exceptions) 2. 20 different R groups C. Classification based on R-group - know one example from each 1. Aliphatic-hydrophobic 2. A r o m a t i c - h y drophobic 3. Polar Uncharged-hydrophilic 4. A c i d i c-hydrophilic 5. B a s i c - hydrophilic V. Polypeptides and Proteins A. Peptide Bond 1. join amino group of one amino acid with carboxyl group of another by forming and amide bond between them ! Peptide Bond 2. C-N bond has partial double bond character B. Peptides and Polypeptides 1. Peptides contain relatively few amino acids linked by peptide bonds: dipeptide, tripeptide, tetrapeptide, …. 2. Polypeptide contains many amino acids and if there are very many amino acids one can call it protein C. Proteins have molecular weights > several thousand and have 3-4 levels of structure 1. Primary Structure (1°) sequence of amino acids connected by peptide bo n d s 2. Secondary Structure (2°) local conformation of peptide bond backbone stabilized by H-bonds: -helix: intrachain H-bonds & -sheet: interchain H-bonds 3. Tertiary Structure (3°): The complete 3-dimensional structure described by the way the polypeptide chain folds back on itself; stabilized by interactions (bonds) between the amino acid R-groups. Hydrophobic Bonds & van der Walls Interactions – most important 4. Quaternary Structure (4°): only some proteins have 4° structure which is the association of more than one polypeptide Complex Polymer (Macromolecule) Polysaccharide (Complex Carbohydrate) Monomer Simple Polymer Monosaccharide (Sugar) Oligosaccharide Nucleotide Oligonucleotide Nucleic Acid Peptid e Polypeptide Protein Amino Acid An Overview of Protein Functions Table 5.1 Describing Macromolecular Structure α carbon α-Amino Acids Amino group Carboxyl group pH ~7 at high pH H +1 Charge - H+ + H+ H O H 3N C C OH R + O H 3N C C O R + 0 Charge - H+ + H+ H At low pH O H 2N C C O R -1 Charge Stereochemistry -- Tetrahedral α-Carbon L-Alanine O O O N C D-Alanine α-Carbon C C C C C O N 20 Different Amino Acids Are Found in Proteins 1-Letter Name 3-Letter 1-Letter Name 3-Letter A Alanine A M Methionine Met C Cysteine Cys N Asparagine Asn D Aspartic Acid Asp P Proline Pro E Glutamic Acid Glu Q Glutamine Gln F Phenylalanine Phe R Arginine Arg G Glycine G S Serine Set H Histidine H T Threonine Thr I Isoleucine Ile V Valine Val K Lysine Lys W Tryptophan Trp L Leucine Leu Y Tyrosine Tyr Fig. 5.16a: Non-polar, hydrophobic aliphatic and aromatic amino acids often cluster together and are found in the interior of proteins Nonpolar side chains; hydrophobic Side chain Glycine (Gly or G) Methionine (Met or M) Alanine (Ala or A) Valine (Val or V) Phenylalanine (Phe or F) Leucine (Leu or L) Tryptophan (Trp or W) Isoleucine (Ile or I) Proline (Pro or P) Fig. 5.16b: Polar uncharged side chains; hydrophilic Serine (Ser or S) Threonine (Thr or T) Cysteine (Cys or C) Tyrosine (Tyr or Y) Asparagine (Asn or N) Glutamine (Gln or Q) Fig. 5.16b: Amino Acids with Hydroxyl Groups in their Sidechains (S, T, Y) These amino acids can also be modified by phosphorylation (addition of phosphate to the hydroxyl group) O- Side chain-O-H Side chain-O-P-OO Fig. 5.16b: Amino Acids with Hydroxyl Groups in their Sidechains (S, T, Y) O-PO3 O-PO3 These amino acids can also be modified by phosphorylation (addition of phosphate to the hydroxyl group) O- Side chain-O-H Side chain-O-P-OO Aspartic acid Glutamic acid (Glu or E) (Asp or D) Lysine (Lys or K) Arginine (Arg or R) Histidine (His or H) Note: similar size and shape but different chemical properties Asparagine (Asn) N Aspartic Acid (Asp) D Glutamine (Gln) Q Glutamic Acid (Glu) E Aromatic side chains (F,W,Y) Tyrosine (Tyr) Y Phenylalanine (Phe) F Tryptophan (Trp) W Ring system in side chain absorbs ultra-violet (UV) light giving us a way of measuring protein concentration Note similar size and shape of Tyr and Phe (only difference is extra –OH group in Tyr making it more hydrophilic) Special cases: Glycine is the smallest amino acid and its small side chain can fit into small spaces in protein Glycine (Gly) G Cysteine (Cys) C The sulfhydryl group (-S-H) of two cysteines can react to form a covalent disulfide bridge (-S-S-) The side chain of proline is covalently linked back to the α-amino group. This limits the rotation of the side chain and introduces kinks in proteins Proline (Pro) P Amino Acids Whose Structures You Need to Know Alanine (Ala) Aliphatic hydrophobic Phenylalanine (Phe) Aromatic hydrophobic Serine (Ser) Polar uncharged Lysine (Lys) Basic Aspartic Acid (Asp) Acidic Fig. 5.17: Peptide Bonds Link Amino Acids Peptide bond New peptide bond forming Side chains Backbone Amino end (N-terminus) Peptide bond Carboxyl end (C-terminus) Peptide Bond: How proteins (polypeptides) are made from amino acids? H H O N C C O H R 2 H H O H 3N C C OH R 1 + H H 2O O O H 3N C C N C C O R1 H R2 + Amide bond Lone electron pair on N forms second bond O C C N C H δ- O Cα C N δ+ Cα H O+ C C N C H or O C C N C H The Peptide Bond group is Polar and Planar (the atoms lie on a plane) The π bond is shared between the O and N in the Peptide Bond Group. Thus, each C=O and C=N bond behaves like a double bond, and there is no rotation around the bonds connecting these atoms. Furthermore, all of the atoms of the peptide bonding group lie on a plane. Peptide Bond: Structural characteristics Peptide Bonds free rotation is not possible around C-O and C-N bonds. Rotation is possible around the single bonds to the Cα’s O C Cα N H Cα H N C Cα O Planar Peptide Bond groups joined at Cα’s Side Chain Main Chain Amino Terminus Carboxy Terminus Aspartame a.k.a. NutraSweet - Is a Dipeptide AspartylAlanineMethylEster, a dipeptide. It is shown in two orientations to demonstrate the 120° bond angles between between the atoms of the peptide bond, and the fact that all of these atoms lie on a plane. 120° Secondary Structure: Local folding of the polypeptide backbone 1.5 Å Hydrogen Bond 3.6 residues/turn 5.4 Å .5 Å Hydrogen Bond Fig. 5.18: Tertiary Structure Describes overall fold of polypeptide backbone β-sheet α-helices Folding puts some amino acid side chains (Hydrophobic) in interior and some (Hydrophilic) on exterior surface of protein Different functional groups on surface give local sites distinct shapes and specific properties Fig. 5.20: What Bonds Stabilize Tertiary Structure? 1. Hydrophobic and van derWaals Interactions: Packing (clustering) of hydrophobic side chains into interior away from water, keeping most hydrophilic side chains on surface. 2. Hydrogen bonds of secondary structure elements 3. Ionic interactions between oppositely charged side chains 4. Some proteins are also stabilized by disulfide bonds between pairs of cysteine side chains Fig. 5.21: The Four Levels of Protein Structure Primary Structure Secondary Structure β pleated sheet +H 3N Amino end Examples of amino acid subunits α helix Tertiary Structure Quaternary Structure Level of Structure Type of Bond Between peptide bond groups Hydrophobic Bond most important + others Fig. 5.22: Changing A Proteins’s Amino Acid Sequence Can Change Its Shape Sickle-cell hemoglobin Normal hemoglobin Primary Structure 1 2 3 4 5 6 7 Secondary and Tertiary Structures β subunit Quaternary Structure Function Normal hemoglobin Molecules do not associate with one another; each carries oxygen. α Red Blood Cell Shape 10 µm β α β 1 2 3 4 5 6 7 Exposed hydrophobic region Sickle-cell hemoglobin α β subunit Molecules crystallize into a fiber; capacity to carry oxygen is reduced. β α β 10 µm Fig. 5.23: Amino Acid (primary structure) Sequence Determines Shape increase temperature, change pH, add chemical agents that disrupt hydrogen bonds, ionic bonds and disulfide bridges Anfinsen experiment (1965) Denaturation (Unfolding) Folding (spontaneous) Normal protein (Biologically active) Renaturation Denatured protein (Biologically inactive) Proteins Form a Variety of Shapes and Sizes http://www.sci.sdsu.edu/TFrey/ProtStructClass/ Quaternary Structure: Some proteins form stable oligomeric structures containing two or more polypeptides Antibodies Hemoglobin Photosynthetic Reaction Center (membrane protein) http://www.sci.sdsu.edu/TFrey/ProtStructClass/ Membrane Proteins: Some are a single polypeptide others have Quaternary Structure Bacteriorhodopsin Photosynthetic Reaction Center Bacterial Porin (membrane protein) http://www.sci.sdsu.edu/TFrey/ProtStructClass/ Cofactors: Some proteins bind ions and/or organic molecules to help them fulfill their function Hemoglobin Myoglobin http://www.sci.sdsu.edu/TFrey/ProtStructClass/ Molecules that interact stably have complementary shapes (fit like a lock-and-key or a hand in a glove) so that they can make lots of weak intermolecular bonds