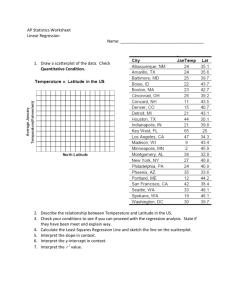
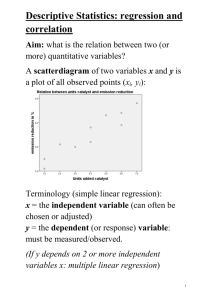
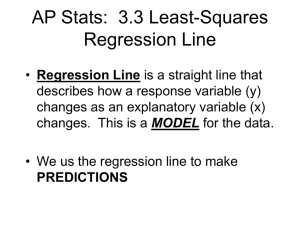
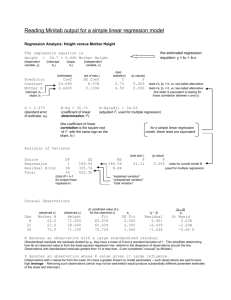
Lecture 2
advertisement

Lecture Two: A Quick Review/Overview on Generalized
Linear Models (GLMs)
Outline
The topics to be covered include:
• Model Definition (Examples, Definition, and Related Topics)
• Parameter Estimation (MLE and WILS Algorithm)
• Model Deviances and Hypothesis Testing Problems (Deviance, Z-test/Wald
Chi-square Test and Deviance Test)
• Residuals and Model Diagnostics (Residuals, Residual Plots, and Measures
of Influence)
1
1
Some Special Examples of GLMs
Logistic Regression Model
• Consider a data set where the response variable takes only 0 or 1 values
(e.g., Yes/No type, we code Yes = 1 and No = 0) and the single covariate
variable is (continues) numerical type.
[Insert Figure 2.1 for an example data]
• If we apply a simple linear regression model
yi = β0 + β1 xi + i
(1)
to fit the data, there are at least three problems from modeling viewpoint:
◦ Problem 1: If model (1) is correct, we will have
Pr(Yi = 1) = E(Yi ) = β0 + β1 xi .
=⇒ This equation is a mismatch! –- because the left hand side can
only take values in interval (0,1) and the right hand side can be any
value between (−∞, ∞).
◦ Problem 2: If model (1) is correct, i = yi − (β0 + β1 xi ). With yi only
taking values 0 or 1, the random error
i can only take two values either (β0 + β1 xi ) or 1 − (β0 + β1 xi ).
=⇒ The assumption that i is normally distributed (or close to be normally distributed) with mean 0 is no longer valid.
◦ Problem 3: If model (1) is correct, the variance of the random error
var(i ) = var(Yi ) = πi (1 − π) = (−ηi )(1 − ηi ).
Here, πi = Pr(Yi = 1) and ηi = β0 + β1 xi .
=⇒ This implies that var(i ) is not a constant, which violates the constant error variance assumption in a simple linear regression model!
Conclusion: It is not appropriate to use the simple linear regression to
model regression data with binary type responses.
• One solution to the mismatch problem is to use the logistic function
g(t) =
2
et
1 + et
to link the linear predictor ηi = β0 + β1 xi to πi the expected value of the
response. Note that, g(t) is a monotonic mapping from (−∞, ∞) to (0, 1).
This is a much better way of specifying the mean model for the response
variable.
[Insert Figure 2.2 here]
• To complete the model specification, we need in addition to specify the
distribution of the random errors (or equivalently, the distribution of the
responses) – which should be different from the normal distributions!
• Indeed, the formal definition of the logistic (regression) model for binary
response data with p covariate is as follows:
A set of regression data {(yi , xi1 , . . . , xip ) : i = 1, 2, . . . , n} with binary type
response yi is said to follow a Logistic regression model, if
◦ The responses yi are independently observed for fixed values of covariates (xi1 , xi2 , . . . , xip )1 , and the covariate variables may only influence the
distribution of the response yi through a single linear function
ηi = β0 + β1 xi1 + . . . + βp xip .
(This linear function is called linear predictor)
◦ The mean of the response πi = E(yi ) is linked to the linear predictor ηi
by the following equation
πi
log
1 − πi
= ηi
or πi =
e ηi
.
1 + e ηi
s
(The function h(s) = log( 1−s
) is called a link function and its inverse
t
e
function g(t) = 1+e
t is called a inverse link function.)
◦ The response yi follows a Bernoulli distribution yi ∼ Bernoulli(πi ).
• Similar as in the regular linear regression, the β0 is known as the intercept
and βk ’s are the slope parameters. One can interpret βk as the expected
amount increase in h(E(y)) with a unit increase in the kth covariate xk ,
when the covariate variable x is a numerical type.
◦ Specifically, in the logistic model,
yi = 1) and h(E(yi )) = log
πi
1−πi
πi
1−πi
is the odds of “success” (i.e.,
is the log-odds. So, in the logistic
1
The covariate variables can be either numerical or categorical and they are considered
as fixed; See, Lecture 1.
3
model, eηi is the odds of the ith response being a success and eβk is the
odds ratio between with and without unit increase in the kth covariate.
Or, in other words, with a unit increase in the kth covariate, we expect
the odds of success to increase eβk times.
[Insert Figure 2.3 here]
• The above logistic model for binary response data can be slightly modified/extended to cover binomial response data: Suppose the ith trial has
ỹi “successes” (1’s) in ni tries. Consider the “success” rate yi = ỹi /ni . The
first two items in the model definition is exactly the same and the third
item is replaced by “The number of successes responses in the ith trial
ỹi = ni yi follow a Binomial distribution ỹi = ni yi ∼ Binomial(ni , πi )”.
[Bring back Figure 2.1 for another example data]
• Since we know the responses yi ’s are from Bernoulli or Binomial distributions, we can write out the likelihood function of observing yi ’s:
L(β|y) =
n
i=1
f (yi ) =
n i=1
ni
πiỹi (1−πi )ni −ỹi = ... = e
ỹi
n
i=1
·
ni {yi ηi −log(1+e´i )}+log
ni
ỹi
We can maximize this likelihood function to obtain the MLE estimates of
unknown regression parameters β = (β0 , β1 , . . . , βp )T , i.e.,
β̂M LE
1
n
n
x
i1
= argmaxβ [ni {yi ηi −log(1+eηi )}], or it solves
ni (yi − πi )
= 0.
...
i=1
i=1
xip
Further details of the estimation along with hypothesis testing and diagnostic will be discussed later.
Poisson Regression Model
• Sometimes one needs to perform regression analysis on Poisson type responses. The possible values of a Poisson type response are 0, 1, 2, 3, . . . .
◦ Some examples:
the number of insurance claims received in a given day by an office
the number of customers served in one day by a sales person.
the number of patients treated in an emergency room each Monday.
4
.
etc.
[Insert Figure 2.4 here]
◦ Similar to in the binary logistic regression case, the regular linear regression model is not appropriate for Poisson type responses (details omitted).
⇒ We need to use a Poisson regression model which relates the response
of Poisson counts to a certain set of covariate variables.
• We can specify the Poisson (regression) model in the same style as we used
to specify the logistic model:
◦ The responses yi are independently observed for fixed values of covariates (xi1 , xi2 , . . . , xip ), and the covariate variables may only influence the
distribution of the response yi through a single linear function
ηi = β0 + β1 xi1 + . . . + βp xip .
(This linear function is called linear predictor)
◦ The mean of the response µi = E(yi ) is linked to the linear predictor ηi
by the following equation
log(µi ) = ηi
or µi = eηi .
(The link function h(s) = log(s) and its inverse function is g(t) = et .)
◦ The response yi follows a Poisson distribution yi ∼ Poisson(µi ).
• We can interpret the slope parameter βk as the expected amount increase
(or change) in the expected value of a response at the (natural) log-scale,
with a unit increase (or change) in the kth covariate.
• We can write out the likelihood function of observing yi ’s:
L(β0 , β1 |y) =
n
i=1
f (yi ) =
n
µyi i e−µi
i=1
yi !
= ... = e
n
i=1
{yi ηi −e´i −log(yi !)}
.
We can maximize this likelihood function to obtain the MLE estimates of
unknown regression parameters β = (β0 , β1 , . . . , βp )T , i.e.,
β̂M LE
1
n
n
x
i1
= argmaxβ (yi ηi − eηi ), or it solves (yi − µi )
= 0.
...
i=1
i=1
xip
5
Further details of the estimation along with hypothesis testing and diagnostic will be discussed later.
Regular Linear Regression with Normal Errors
• Regular Gaussian linear regression model
yi = β0 + β1 xi1 + β2 xi2 + . . . + βp xip + i
where i ∼ N (0, σ 2 ).
This model assumption is the same as the following two requirements:
◦ E(Yi ) = ηi , where ηi = β0 + β1 xi1 + β2 xi2 + . . . + βp xip ;
◦ i ∼ N (0, σ 2 ) (or, equivalently, yi ∼ N (ηi , σ 2 ) (given covariates xi )).
⇒ We can specify the Gaussian linear regression model in the same style
as we used to specify the logistic and Poisson models:
◦ The responses yi are independently observed for fixed values of covariates (xi1 , xi2 , . . . , xip ), and the covariate variables may only influence the
distribution of the response yi through a single linear function
ηi = β0 + β1 xi1 + . . . + βp xip .
(This linear function is called linear predictor)
◦ The mean of the response µi = E(Yi ) is linked to the linear predictor ηi
by the following equation
µi = η i .
(Here, both the link and inverse link functions are identity functions
h(s) = g(t) = t.)
◦ The response yi follows a normal distribution yi ∼ N (µi , σ 2 ).
6
2
Formal Definition of GLMs
Formal Definition of GLMs
• Generalized linear models (GLMs) extent linear regression models to accommodate both non-normal response distributions and transformation of
linearity. A formal definition is provided as follows.
A regression data set containing responses yi and covariates xi is said to
follow a generalized linear model (GLM) is
◦ The responses yi are independently observed for fixed values of covariates (xi1 , xi2 , . . . , xip ), and the covariate variables may only influence the
distribution of the response yi through a single linear function
ηi = β0 + β1 xi1 + . . . + βp xip .
(This linear function is called linear predictor)
◦ The mean of the response µi = E(yi ) is linked to the linear predictor ηi
by a smooth invertible link function
h(µi ) = ηi .
(Here, h(s) is called a link function and its inverse function g(t) = h−1 (t)
is called the inverse link function.)
◦ The distribution of the response yi has density of form
Ai
φ
f (yi |β, φ) = exp
{yi θi − γ(θi )} + τ (yi , ) ,
φ
Ai
(2)
where φ is a scale parameter 2 that could be either known or unknown,
Ai is a known constant, and θi = θ(ηi ) is a function of linear predictor
ηi .
• Remark 1: We can prove that the function θ(·) = (γ )−1 (g(·)). So it is
completely determined by the function γ and the link function h.
• Remark 2: GLM is fully determined by the choices of the link function h
and the form of response distribution (i.e., the form of the function γ).
2
Some books (for example, McCullaugh and Nelder, 1989) also call φ a dispersion
parameter.
7
• Remark 3: It is easy to show that
E(yi ) = µi = γ (θi ) and var(yi ) =
φ γ (θi ).
Ai
• Remark 4: We can interpret the slope parameter βk as the expected amount
increase (or change) in h(E(y)) with a unit increase (or change) in the kth
covariate.
Verifying Some Special Examples:
• Gaussian/Normal For the normal distribution, we can write φ = σ 2 and
1
1
1
1
f (yi ) = exp {yi µi − µ2i − yi2 } − log(2πφ) .
φ
2
2
2
So, θi = µi = ηi and γ(θi ) = θi2 /2.
• Binomial (logistic) For a binomial distribution, we take φ = 1 and write
·
f (yi ) = exp ni {yi ηi − log(1 + eηi )} + log
ni
ni yi
So, Ai = ni , θi = ηi and the function γ(θi ) = log(1 + θi ).
• Poisson For a Poisson model, we take φ = 1 and write
f (yi ) = exp{(yi ηi − eηi ) − log(yi !)}
So, θi = ηi , and γ(θ) = eθi .
• Gamma model is used to model Gamma-distributed responses where the
response variance var(yi ) ∝ µ2i with µi = E(yi ).
◦ In a Gamma model, the mean µi is often link to the linear predictor ηi
by a inverse link function h(t) = 1/t:
µi = ηi−1 .
◦ Also, in the Gamma model, the response yi follows a Gamma distribution with density
1
f (yi ) =
Γ(α)
8
α
µi
α
yiα−1 e
−
®yi
¹i
,
where E(yi ) = µi and var(yi ) = µ2i /α. This density function can be
written as
f (yi ) = exp [α{yi ηi − log(ηi )} + (α − 1) log(yi ) − α log(α) − log{Γ(α)}] .
So, φ = 1/α, θi = ηi , and the function γ(θi ) = log(θi ).
Link Functions and Model Family
• Some commonly used link functions:
◦ Logit
t
h(t) = log
1−t
where Φ(t) =
◦ Probit
h(t) = Φ−1 (t),
t
−∞
1 2
1
√ e− 2 z dz
2π
◦ Complementary log-log
h(t) = log{− log(1 − t)}
◦ Identify
h(t) = t
◦ log-link
h(t) = log(t)
◦ Square root
h(t) =
√
t
◦ Inverse
h(t) = 1/t
• The combination of the response distribution and the link function is called
the family of GLM.
◦ See Figure 2.5 for the usual combination.
[Insert Figure 2.5]
9
• Canonical link function For each response distribution, there is a choice of
link function such that θi = ηi is always true in the form (2). Such a choice
of link function is called the canonical link function.
◦ The choice of the canonical link is mathematically convenient and the
minimal sufficient statistic for β can be easily obtained.
◦ The default choices of the link functions are usually the canonical links.
[Insert Figure 2.5 here]
10
3
Parameter Estimation in GLMs
MLE and Likelihood Estimating Equations
• In GLM, the unknown regression parameters β are estimated by the maximum likelihood estimators β̂ = β̂MLE . That is, β̂ maximizes the (log-)
likelihood function
n
!(β|y) =
i=1
Ai
φ
{yi θi − γ(θi )} + τ (yi , ) .
φ
Ai
• Taking derivative on !(β|y) with respective to β and set them equal to
0, the estimators β̂ are the solution to the following likelihood estimation
equations:
1
n
(yi − µi )g (ηi )
xi1
=0
(3)
...
var(yi )
i=1
xip
◦ Recall 1: In the logistic regression, we solve equations
1
n
x
i1
ni (yi − πi )
= 0.
...
i=1
xip
(Note: var(yi ) = πi (1−πi )/ni , πi = E(yi ) = µi , and g (ηi ) = πi (1−πi ).)
Recall 2: In the Poisson regression model, we solve equations
1
n
x
i1
(yi − µi )
= 0.
...
i=1
xip
(Note: var(yi ) = µi and g (ηi ) = eηi = µi .)
WILS algorithm
We use a so called weighted iterative least square (WILS) algorithm to solve
the estimating equations (3).
11
• The working response for the ith observation in GLM setting is defined by
zi = η i +
(yi − µi )
dηi
=
η
+
(y
−
µ
)
.
i
i
i
g (ηi )
dµi
It behaves in many ways like the regular response yi in the regular linear
regression model!
◦ The weight for the ith observation is
wi =
[g (ηi )]2
var(yi )
• In terms of zi , the likelihood estimating equations (1) can be re-expressed
in a form that very much similar to the regular LS estimating equations:
1
n
x
i1
= 0 or, in matrix form XT WXβ = XT Wz.
wi (zi − ηi )
...
i=1
xip
Here, X is the regular design matrix (the same as in linear regression),
z = (z1 , z2 , . . . , zn )T and W is a diagonal matrix with diagonal entries
equal to w1 , w2 , . . . , wn , respectively.
• The above estimating equations lead us to the WILS algorithm:
◦ Step 0: Find a set of staring values, say β (0) .
◦ Step 1: For m = 0, 1, 2, ..., compute the current working responses
and the weights by replacing the unknown parameters with their current
estimates, i.e., z(m) = z| β=β (m) and W(m) = W| β=β (m) . Then, update the
unknown parameter estimates by the weighted least formula:
β (m+1) = {XT W(m) X}−1 XT W(m) z(m) .
◦ Step 2: Repeat Step 1 until the updated estimates β (m+1) and current
estimates β (m) are very close (convergence).
(Under very mild conditions, the WILS converges.)
Some Theoretical Results
12
• Under very mild conditions, when n is large, the distribution of the estimators β̂ is asymptotically a normal distribution N (β, In−1 ).
◦ Here, In =
matrix.
∂2
∂β∂β T
!(β|y) = ... = XT WX is the Fisher information
• This theorem implies:
◦ The β̂ are consistent estimators (when n large, they are close to the
corresponding true parameter values).
◦ The β̂ are asymptotically efficient estimators and a way to estimate the
variance of the estimators β̂ is by the Fisher information matrix.
13
4
Model Deviance and Hypothesis Testing Problems
Model Deviance
• (Overall statement) Model deviance in GLM setting is an analogous concept
to the residual sum of squares (SSE) in the regular linear regression model.
It is a key quantity that is involved in many problems of hypothesis testing
and model diagnostics.
• (Recall) the linear regression model with normal errors,
yi = µi + i ,
where µi = β0 + β1 xi1 + . . . + βp xip and i ∼ N (0, σ 2 ).
The log likelihood function in terms of responses yi ’s and their means µi ’s
is
n
1 n
l(µ|y) = − log(2πσ 2 ) − 2
(yi − µi )2 .
2
2σ i=1
• The saturated model in GLM setting refers to the perfectly fitting (i.e.,
over-fitting) model of using n free parameters to fit n observations. In a
saturated model, the log-likelihood function achieves its maximum achievable value.
◦ For example, in the normal linear regression case, we have µ̃i = ŷi = yi
and this leads to l(y|y) = − n2 log(2πσ 2 ) which is an upper-bound of the
likelihood functions.
• The residual sum of squares SSE = ni=1 (yi − ŷi )2 is a quantity used to
measure the discrepancy (variation) between the fitted model values and
the observed data, and its format comes from the squared error loss. We’d
like to point out that SSE can also be expressed in terms of twice the
discrepancy of the likelihood functions between the currently fitted model
and the saturated model:
SSE = 2σ 2 {l(y|y) − l(µ̂|y)} = . . . =
n
(yi − µ̂i )2 .
i=1
• Model deviance in GLM is defined as the twice discrepancy of the loglikelihood functions of the fitted model and the saturated model. In math14
ematical form, the model deviance is defined as
n
DM = 2φ{l(y|y) − l(µ̂|y)} = 2
Ai {yi θ̃i − γ(θ̃i )} − {yi θ̂i − γ(θ̂i )} ,
i=1
where θ̃i = (γ )−1 (µ̃i ) with µ̃i = yi and θ̂i = (γ )−1 (µ̂i ) with µ̂i = g(η̂i ) and
η̂i = β̂0 + β̂1 xi1 + . . . + β̂p xip .
• When the scale parameter φ in the GLM definition is not 1, we also define
the scaled model deviance, which is the scaled version of the model deviance:
∗
=
DM
n
DM
Ai {yi θ̃i − γ(θ̃i )} − {yi θ̂i − γ(θ̂i )} .
= 2{l(y|y) − l(µ|y)} = 2
φ
i=1 φ
• In the binomial logistic regression model, the log-likelihood function can
be expressed as
n
l(µ|y) =
i=1
µi
ni
ni {yi log
+ log(1 − µi )} + log
ỹi
1 − µi
and the scale parameter φ = 1. So, the model deviances are
n
∗
=2
DM = DM
{ni log(
i=1
where µ̂i = π̂i =
e´ˆi
1+e´ˆi
yi
1 − yi
) + ni (1 − yi ) log(
)},
µ̂i
1 − µ̂i
and η̂i = β̂0 + β̂1 xi1 + . . . + β̂p xip .
• In the Poisson model, the log-likelihood function can be expressed as
n
{yi log(µi ) − µi − log(yi !)}
l(µ|y) =
i=1
and the scale parameter φ = 1. So, the model deviances are
∗
DM = DM
=2
n
{yi log(yi /µ̂i ) − (yi − µ̂i )} .
i=1
Here µ̂i = eη̂i and η̂i = β̂0 + β̂1 xi1 + . . . + β̂p xip .
15
• Unfortunately, unlike SSE ∼ σ 2 χ2n−p−1 in the linear regression case, DM
in general does not follow a χ2 distribution, even asymptotically! Neither
does the generalized Pearson chi-square statistic, which is defined by
n
X2 =
(yi − µi )2
.
i=1 var(µ̂i )
Note, For the normal distribution, both X 2 and DM are the residual sum
of squares (SSE), while for the Poisson and Binomial models, X 2 is the
original Pearson Chi-square statistic.
• The good news is that the difference of the deviance functions between
two nested models does follow a χ2 distribution when n is large. (This is
analogous to SSE(R) − SSE(F ) ∼ σ 2 χ2p−q .) This result will be used to
test between two nested models below.
Hypothesis Testings
• Z-test for the jth regression parameter βj . In GLM, we also may need to
test the hypothesis whether the jth covariate has significant contribution
to response variable or not. In math term, the hypotheses are H0 : βj = 0
versus H1 : βj = 0. Recall that the maximum likelihood β̂ is asymptotically
normally distributed. To formally conduct the test, we use the z-statistic
Zj = se(β̂β̂j ) and compare it to the standard normal distribution.3 When the
j
absolute value |Zj | is large (corresponding to small p-value), we reject the
null hypothesis H0 : βj = 0.
◦ Some software reports Zj2 , which is compared to χ21 -distribution. It is
known as the Wald chi-square test and it is equivalent to the Z-test.
• Deviance chi-square test for nested models. To answer the question whether
a subset of the regression parameters are significant or not in a linear regression model, we use the deviance chi-square test for nested models. This test
is analogous to the F -test for nested models in the regular linear regressions.
Without loss of generality, assume that we want to test whether the last
p − q (q < p) parameters are zero or not, that is, H0 : βq+1 = . . . = βp = 0
versus H1 : One of these p − q parameters are not equal to 0. Under the
null hypothesis H0 , the regression model has q covariates and we call this
3
Note, when n → ∞, the tn−p -test in the normal linear regression model is the same
as z-test.
16
model a reduce model (R); the model under H1 has p covariates and we call
it a full model (F). To formally conduct the test, we use the test statistic
that is the difference of model deviances between these two nest model:
DM (R) − DM (F )
.
φ
When φ is known, this test statistic is compared to the χ2p−q -distribution
with degrees of freedom p − q. If this statistic is large (corresponding to
p-value small), we reject the null hypothesis H0 : βq+1 = . . . = βp = 0.
When φ is not known, we need to replace φ by its estimator φ̂ and still
compare it to the χ2p−q -distribution.4,5
• Similar to ANOVA tables in linear regression, the results in the aforementioned deviance chi-square test for nested models are summarized in a
table. Such a table is known as the analysis of deviance table.
• Goodness of ¯t tests. In general, goodness of fit test for GLM is not
available, unless in special cases.
• One commonly used goodness of fit test in logistic regression model is the
Hosmer-Lemeshow goodness of ¯t test.
◦ The test can be described as this: we place the n observations into approximate 10 groups (sorted/grouped according to their estimated probablities of successes) and calculate the corresponding generalized Pearson
chi-square statistic. To test the model, this Pearson chi-square statistic
(after grouping) is compared to a chi-square distribution.
◦ The basis for this Homser-Lemeshow test: asymptotic chi-square distribution holds for DM (also DM ) in a binomial model if, as n increases,
the number of covariate cells (groups) are fixed and the observations in
each cell increases.
• Similar grouping technique can be used to constructe a goodness of fit test
for Poisson models.
4
Depending on how the scale parameter φ is defined, sometimes we can use an F test
instead of the Chi-square test.
5
The scale parameter φ can be estimated either by its moment estimators X 2 /(n−p−1)
or DM /(n − p − 1), or by its mximum likelihood estimatior.
17
5
Model Diagnostics
Residuals
• The most intuitive way to define a residual is to define the response residual
= g(η̂ ).
where µ̂i = Ey
i
i
riR = yi − µ̂i ,
Although it is intuitive but loses a lot of nice features known associated
with residuals in the regular linear regression models. We usually do not
use it in model diagnostics.
• Deviance residuals. The nice feature of the deviance residuals is that they
are tied to the likelihood function and model deviance, thus the model
fitting. The deviance residual for the ith observation is defined as
riD = ai di ,
where di is the contribution of the ith observation to the model deviance
and ai = 1 if yi > µ̂i , ai = 0 if yi = µ̂i and ai = −1 if yi < µ̂i . It is easy to
see that
n
n
DM =
(riD )2 .
di =
i=1
i=1
This equation is comparable to the equation SSE =
linear regression model.
n
2
i=1 ei
in the regular
◦ In the binary logistic regression model, the deviance residual is
riD
2{log(1 + eη̂i ) − η̂i },
= − 2 log(1 + eη̂i ),
if yi = 1;
if yi = 0;
where η̂i = β̂0 + β̂1 xi1 + . . . + β̂p xip .
◦ In Poisson models, the deviance residual formula is
riD
2{yi log(yi /µ̂i ) − yi + µ̂i },
= − 2{y log(y /µ̂ ) − y + µ̂ },
i
i
i
i
i
if yi − µ̂i > 0;
if yi − µ̂ < 0;
where µ̂i = eη̂i and η̂i = β̂0 + β̂1 xi1 + . . . + β̂p xip .
18
• Working residuals. Recall that the working responses zi defined in WILS
algorithm. We mentioned that it works like the ith response in the regular
linear regression model. The working residual for the ith observation is
defined as:
riW = (zi − ηi )| β=β̂ = (yi − µ̂i )
∂ηi
| β=β̂ = (yi − µ̂i )/g (η̂i ).
∂µi
The working residual is easily available from WILS algorithm and they are
tied to the generalized Pearson chi-square statistic.
◦ In logistic regression model, the working residual is
(yi − µ̂i )
,
µ̂i (1 − µ̂i )
riW =
where µ̂i =
e´ˆi
1+e´ˆi
and η̂i = β̂0 + β̂1 xi1 + . . . + β̂p xip .
◦ In Poisson models, the working residual formula is
riW =
(yi − µ̂i )
,
µ̂i
where µ̂i = eη̂i and η̂i = β̂0 + β̂1 xi1 + . . . + β̂p xip .
• The Pearson residual is closely related to the Pearson statistic X 2 :
yi − µi
.
riP = var(µ̂i )
Clearly, X 2 = ni=1 (riP )2 . Pearson residuals are rescaled version of the
working residuals, when proper account is taken of the associated weights
√
riP = wi riW . Here, wi is defined in the WILS algorithm.
◦ In logistic regression model, the working residual is
(yi − µ̂i )
riP = ,
µ̂i (1 − µ̂i )
where µ̂i =
e´ˆi
1+e´ˆi
and η̂i = β̂0 + β̂1 xi1 + . . . + β̂p xip .
19
◦ In Poisson models, the working residual formula is
riW =
(yi − µ̂i )
√
,
µ̂i
where µ̂i = eη̂i and η̂i = β̂0 + β̂1 xi1 + . . . + β̂p xip .
• The most used residuals in model diagnostics are the deviance residuals.
Then, followed the working residuals and the Pearson residuals. Other
versions of residual definitions are also available, for example, the Anscombe
residual tries to make the residuals “as close to normal as possible”.
• A version of deletion residual is also available under GLM setting, which is
related to the Pearson residual. Their forms are very complicated and we
are thus omitting it here.
Graphical Methods
In the GLM setting, we can still plot the regular residual plots, where
deviance residuals (or working residuals or Pearson residuals) are plotted
against the jth covariate values xij , or the fitted values of the linear predictor η̂i ’s, or other things. But the interpretation is less clear and patterns
may vary quite differently for different models. In practice, we often turn to
other specialized residual plots and try to detect any systematic departure
from the fitted model. Here we discuss three commonly used residual plots
in GLMs.
• Index plot of deviance residuals plots the deviance residual riD against
the index of observation i, and then use a straight line to connect the
neighboring points. This plot helps to identify outlying residuals (but
they do not indicate whether the outlying residuals should be treated as
outliers).
[Insert Figure 2.6 here]
• Half normal plot in GLM setting is an extension of Atkinson’s (1985) half
normal plot in regular linear regression models. In the plot the absolute
values of the deviance residuals riD are ordered and the kth largest absolute
residual is plotted against z( k+n−1/8
). Here, z(α) is the α-percentile of the
2n+1/2
standard normal distribution. As in the linear regression model, the half
normal plot in GLM setting can also be used to detect influential points.
20
◦ To identify outlying residuals, we also can use the simulated envelope
as in the linear regression models. This envelope constitutes a band such
that the plotted residuals are likely to fall within the band if the fitted
model is correct. Some details of the simulation in the binary logistic
model are
Step 1. For each of the n cases, generate a new Bernoulli outcome yi = 0 or 1
with the success rate πi equal to the estimated probability of response yi = 1.
Fit a logistic regression to the new set of data of size n, with the covariates
keeping their original values. Order the absolute deletion residuals in ascending
order.
Step 2. Repeat step 1 eighteen times (total 19 times).
Step 3. For each k, k = 1, 2, . . . , n, assemble the kth smallest absolute residuals from the 19 groups and determine the minimum value, the mean, and the
maximum value of these 19 kth smallest absolute residuals.
Step 4. Plot these minimum, mean, and maximum values against z( k+n−1/8
2n+1/2 )
on the half-normal probability plot for the original data and connect the points
by straight lines.
[Insert Figure 2.7 here]
• As in the linear regression model, Partial residual plot is useful for identifying the nature of relationship for an independent variable under consideration, say the kth covariate x̃k = (x1k , x2k , . . . , xnk )T , for addition to
the regression model. The partial residuals for the kth covariate in a GLM
model is defined as:
[k]
ri = riW + β̂k xik
for i = 1, 2, . . . , n,
where riW is the working residual for the ith observation. The partial
[k]
residual plot for the kth covariate plots ri against xik . If the response
h(E(yi )) is linearly related to the kth covariate xik , the points should be
more or less around a straight line.
[Insert Figure 2.8 here]
Diagnostics for High Leverage Points
• We can still use the hat matrix to detect the high leverage point. But in
21
1
1
GLM models, the hat matrix is modified as H = W 2 X(XT WX)−1 XT W 2 ,
where W is as defined in the WILS algorithm.
• Let hii be the ith diagonal element of this hat matrix H. Again, we call
those observations with very large hii values high leverage points. We use
the same heuristic rule: any observation with its corresponding hii > 2(p+1)
n
is considered as a high leverage point.
• Note that for GLMs a point at the extreme of the x-range will not necessarily have high leverage if its weight is very small.
Measures of Influence
Many measures of influence described for linear regression models have analogies and are appropriate for the GLM models (especially in binary and
Poisson models). These include DFBETAS, DFFITS, and Cooks Distances,
among others.
• DF BET AS measures the effect of deleting a single observation (say, ith
observation) on the estimator of a particular regression parameter (say, kth
parameter βk ).
β̂k − β̂k(i)
(DF BET AS)k(i) =
,
√
s(i) ckk
where ckk is the kth diagonal entry of (XT WX)−1 .
◦ The Interpretation is that if this is large, then the ith observation has
an undue influence on the kth parameter estimate.
◦ Heuristic rule: to flag any
√ value of |DF BET AS| that is bigger than 1
for a small data set or 2/ n for a large data set.
• Cook's Distance is intended again in GLMs as an overall measure of the
influence of the ith observation on all parameter estimates:
Di =
(β̂ − β̂(i) )T XT WX(β̂ − β̂(i) )
(p + 1)φ̂
.
◦ We may still use the same heuristic rule, which suggests to identify the
ith observation as influential if Di is great that the 10% quantile of the
Fp+1,n−(p+1) distribution, and highly influential if it is greater that 50%
quantile of the Fp+1,n−(p+1) distribution.
22
Other Topics (not going to details)
• Transformations of variables on the covariates (e.g. Box-Tidwell transformation)
• Model selection criteria (e.g., Cp , AIC, BIC) and Stepwise Regressions
6
References
McCullagh, P. and Nelder, J.A. (1989). Generalized Linear Models. 2nd
edition. Chapman and Hall, New York.
23