What makes me tick…tock? June 2012 Lesson 3: How can genetics
advertisement
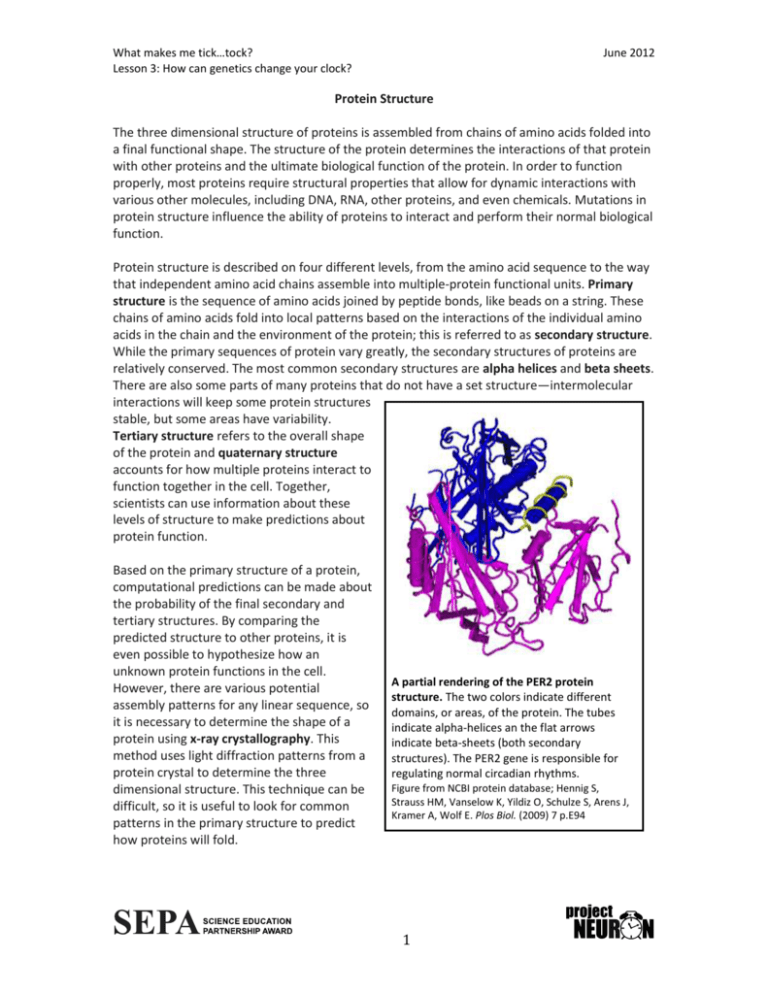
What makes me tick…tock? Lesson 3: How can genetics change your clock? June 2012 Protein Structure The three dimensional structure of proteins is assembled from chains of amino acids folded into a final functional shape. The structure of the protein determines the interactions of that protein with other proteins and the ultimate biological function of the protein. In order to function properly, most proteins require structural properties that allow for dynamic interactions with various other molecules, including DNA, RNA, other proteins, and even chemicals. Mutations in protein structure influence the ability of proteins to interact and perform their normal biological function. Protein structure is described on four different levels, from the amino acid sequence to the way that independent amino acid chains assemble into multiple-protein functional units. Primary structure is the sequence of amino acids joined by peptide bonds, like beads on a string. These chains of amino acids fold into local patterns based on the interactions of the individual amino acids in the chain and the environment of the protein; this is referred to as secondary structure. While the primary sequences of protein vary greatly, the secondary structures of proteins are relatively conserved. The most common secondary structures are alpha helices and beta sheets. There are also some parts of many proteins that do not have a set structure—intermolecular interactions will keep some protein structures stable, but some areas have variability. Tertiary structure refers to the overall shape of the protein and quaternary structure accounts for how multiple proteins interact to function together in the cell. Together, scientists can use information about these levels of structure to make predictions about protein function. Based on the primary structure of a protein, computational predictions can be made about the probability of the final secondary and tertiary structures. By comparing the predicted structure to other proteins, it is even possible to hypothesize how an unknown protein functions in the cell. However, there are various potential assembly patterns for any linear sequence, so it is necessary to determine the shape of a protein using x-ray crystallography. This method uses light diffraction patterns from a protein crystal to determine the three dimensional structure. This technique can be difficult, so it is useful to look for common patterns in the primary structure to predict how proteins will fold. A partial rendering of the PER2 protein structure. The two colors indicate different domains, or areas, of the protein. The tubes indicate alpha-helices an the flat arrows indicate beta-sheets (both secondary structures). The PER2 gene is responsible for regulating normal circadian rhythms. Figure from NCBI protein database; Hennig S, Strauss HM, Vanselow K, Yildiz O, Schulze S, Arens J, Kramer A, Wolf E. Plos Biol. (2009) 7 p.E94 1











