Restriction mapping by double digestion
advertisement

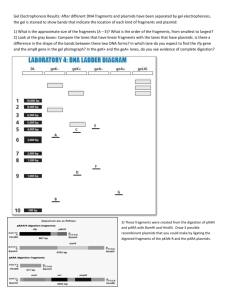
Context: Restriction mapping of clones by double digestion was a major activity of the 1980's and 1990's. Its purpose was to provide a framework of map positions within which to locate genes and other targets of interest. The fragments corresponding to the targets of interest could then be isolated from a gel based on their known sizes and subjected to further characterization. As sequencing technology improved, it became more common to sequence first and then determine the restriction map and location of targets of interest directly from the sequence. Hence the kinds of mapping problems exemplified below where there is no sequence data to assist are now not commonly encountered. An example of mapping by double digestion: This is a plasmid, i.e. a circle. Step 1) Measure single and double digestion product sizes, preferably with lanes side-by-side as indicated below (in Kb): B 2.0 0.76 0.62 B/RI RI R/H 1.8 0.74 0.40 0.20 0.15 2.1 1.3 2.0 0.81 0.30 0.19 H 2.9 0.29 0.20 H/B B 1.8 0.6 0.4 0.21 0.15 2.0 0.76 0.62 Step 2) Evaluate accuracy; in this case let's say we have a 5% measuring error. Step 3) Add up sizes and number of fragments for each digest B 3.38 (3) B/RI 3.29 (5) RI 3.4 (2) R/H 3.30 (4) H 3.39 (3) H/B 3.16 (5) B 3.38 (3) Step 4) Account for missing fragments (doublets or fragments too small to detect. For a circular starting material, the number of double digested bands should = the sum of the single digested bands. (For a linear DNA, the number of double digest fragments would be one less). So we see that the R/H digest is missing one band, and the H/B digest is missing one band. The ave. of the sum of bands in lanes that are not missing fragments is 3.37. 5% error would allow a deviation of 0.16 by accumulated measuring error. The missing lengths for R/H and H/B are 0.07 and 0.21 respectively. Within error, either of these could be too small to see, or a doublet with one of the smaller bands on the gel. In this case, we confirm that the H/B bands is a doublet, because it is twice as bright as it should be. There is no bright band on the R/H lane, so we just call the missing fragment s for "too small to detect". Here are the corrected sizes: B 2.0 0.76 0.62 B/RI RI R/H 1.8 0.74 0.40 0.20 0.15 2.1 1.3 2.0 0.81 0.30 0.19 s H 2.9 0.29 0.20 H/B B 1.8 0.6 0.4 0.21 0.15 0.15 2.0 0.76 0.62 Step 5) Identify the bands coming from the vector (if possible). In this case the vector is sequenced and should have the following map: The HindIII and RI sites are part of a multiple cloning region, and are very close together. Step 6) Set up the boundaries of the insert in the following format: Step 7) Starting with the largest double digest fragments, write down the combinations of doubles that can fit into each of the singles. Keep in mind the limitations imposed by the sizing uncertainty, the vector information, and the rule that each double digest fragment must appear only once in each single digest. B/R 1.8 can only fit into R 2.1. Sizing error would allow a number of combinations of small B/R fragments to fill out the R 2.1, but the vector information says that R 2.1 = B/R 1.8 + 0.2. All the B/R fragments left must fit into R 1.3, so R 1.3 = B/R 0.74 + 0.4 + 0.15. A number of other combinations would be possible by sizing alone. Similarly for Bam: B/R 1.8 can only fit into B 2.0, but there are several combinations of small B/R fragments that could fill out B 2.0. B/R 0.74 can not fit into B 2.0 because we have already established that B/R 1.8 is in there. So only B 0.76 is big enough to contain B/R 0.74. There is not enough room for any smaller B/R fragment in B 0.76, so B 0.76 is B/R 0.74. The remaining B/R fragments must fit into B 0.62. Sizing error only permits the combination of B/R 0.2 and 0.4. That reduces the possibilities for B 2.0 to just B/R 1.8 + 0.15. The object of step 7 is to reduce each of the single digest fragments to a unique combination of double digest fragments. If this can not be accomplished, at least for most of the larger single digest fragments, then additional information will be required to complete the map. Step 8) Fill out the map in a stepwise fashion as follows: R2.1 = 1.8+0.20 R1.3 = 0.74+0.4+0.15 B2.0 = 1.8+0.15 B7.6 = 7.4 B0.62= 0.2+0.4 R1.3 = 0.74+0.4+0.15 B0.76 = 0.74 B0.62 = 0.4+0.2 Notice a very useful mapping relationship demonstrated by the packing of 3 double digest fragments into R1.3. When more than two fragments pack into a single, the 2 on the ends must be unique to the double digest, and the ones in the middle must be identical to fragments in the other single digest. Step 9) Complete the map for R/H and B/H and see that they confirm the R/B map. Example 2: This is a bacteriophage lambda clone. The number of fragments is enough to make mapping by double digestion difficult. This problem includes some southern blotting to help clarify the map. Use of Southern blotting to clarify complex maps. When mapping a large insert, there are usually too many fragments to work from sizes alone. There are many ways to generate additional constraints. Here is a mapping problem involving an insert in a lambda vector, where we will see how Southern blot hybridization can help solve the map. The answer is shown first to help you visualize how the experiment works. The single and double digest fragments are probed with a cDNA (*), a repetitive sequence (&), and the lambda vector ($). R 14.0 10.4 10.0 8.7 7.1 1.6 $ * $ & * & R/B 14.0 10.0 6.0 4.8 4.0 3.2 3.1 2.0 1.9 1.6 1.2 $ $ * B 18.0 11.9 6.3 6.0 4.8 4.8 $ $ * * & * & & When assembling the combinations of double digest fragments, we have the additional constraint that the single containing a double must hybridize to the same probes that the double does. $14 can only be $R14 $10 can only be $R10 *6.0 must go into one of *R10.4 and *R7.1, and *3.1 must go into the other. &2.0 will only fit into &R8.7, so &1.6 must be &R1.6. 4.8, 4.0, 3.2, and 1.9 must go into the remaining space of *10.4, &R8.7, and *R7.1. Look for cases where 3 or more doubles must pack into one single. None of the doubles are big enough to fill out &R8.7, so &R8.7 must have a third fragment. Remembering the rule that if three doubles fit into a single, one of the doubles must be identical to a single in the other single digest, only 4.8 is a candidate to go into &R8.7; so &R8.7 can only be &2.0+4.8+1.9 Similarly either *R10.4 or *R7.1 must contain 3 doubles to account for all the remaining doubles, and *6.0 is the only candidate for the third fragment in a combination. That leaves the only available assignments to be: *R10.4 = 3.2 + *6.0 + 1.2 *R7.1 = *3.1 + 4.0 Similarly for Bam $14 + ? must go into $B18 $10 + ? must go into $B11.9 &2.0 + &1.6 + 1.2 must go into &4.8 If you have > 5% accuracy then *6.0 must be *B6.0 and *3.1 + 3.2 must be *B6.3 If you don't have > 5% accuracy you'll have to treat these two Bam fragments as a doublet (*B6.0a & *B6.0b) and admit that you can't tell which one is = *6.0 and which one is = *3.1 + 3.2 4.8 we've already established is = B4.8 4.0 has no place to go but into $B18, and 1.9 has no place to go but into $B11.9. Now that all doubles are uniquely assigned, the map can be assembled as before. Genomic Southern blot Unique fragments purified or cloned from the insert should detect the same size fragments when used as a probe in a Southern blot of the source genome as they do in the clone. If not, beware of a double insert.



![[#SWF-809] Add support for on bind and on validate](http://s3.studylib.net/store/data/007337359_1-f9f0d6750e6a494ec2c19e8544db36bc-300x300.png)