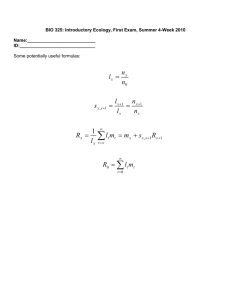
Supplemental data-Immuno-electron microscopy and 3d
advertisement

Supplementary Information: Acute imaging 0 hour 1 hour Supplementary Figure 1. Acute Imaging. Example of stable spines over 1 hour (e.g. yellow arrowhead), and a new thin spine that appeared (blue arrow). Scale bar, 5μm. Acute experiments were performed in anesthetized young adult mice (PND 35-50; n=4). Images were collected every 15 minutes for 2 hours. Relatively few changes occur over these time periods (~2.7% / hour, 367 spines; Supplementary Fig. 1 a, b). Therefore daily imaging captures the majority of the spine dynamics. Supplementary Information : Immuno-electron microscopy and 3d reconstruction Imaged GFP-labeled dendrites were localized at the ultrastructural level using preembedding immunocytochemistry. Within 10 minutes of the final in vivo imaging session, the already anaesthetized mice were transcardially perfused with a solution of 0.2% glutaraldehyde and 2% paraformaldehyde in 0.1M phosphate buffer, pH7.4. One hour after perfusion, brains were removed and 60m vibratome (Leica VT100) serial sections were cut tangential to the surface of barrel cortex, approximately parallel to the optical sections collected during the in vivo imaging. These sections were then cryoprotected, freeze thawed in liquid nitrogen, then pretreated in 0.3% hydrogen peroxide in PBS. After blocking in PBS containing 5% goat serum and 0.1% bovine serum albumin-c (Aurion, Netherlands), they were incubated overnight in primary antibody (rabbit anti-GFP, Chemicon #AB3080) at 4C, then in biotinylated secondary antibody (1:500 goat anti-rabbit (F)ab fragment, Jackson Laboratories, USA) for 4 hours before being incubated in avidin biotin peroxidase complex (ABC Elite, Vector Laboratories, USA), and finally enhanced in 3, 3’-diaminobenzidine tetrachloride and 0.015% hydrogen peroxide. Sections were postfixed in 1% osmium tetroxide in cacodylate buffer (0.1M), dehydrated in a graded series of alcohol followed by propylene oxide and then embedded between silicon coated glass slides in Epon resin (Fluka). Once cured, the resin embedded sections were viewed under a light microscope and compared to projections of image stacks collected in vivo (Fig. 3 a-b). This was done by matching an image of the vascular pattern, taken immediately after the final in vivo imaging session, with the arrangement of blood vessels seen in the first few tangential sections. The exact position of particular dendrites could then be identified in relation to the pattern of vessels on the surface of the cortex. A total of 1,534 serial thin sections (silver/gray interference color) were cut from this region and placed in formvar-coated, single slot grids. Images of labeled dendrites were recorded at a magnification of 13,500 times using a Phillips CM10 transmission electron microscope at a filament voltage of 80kV and collected digitally using a CCD camera (Gatan, USA) (see Supplementary Movie showing sequential serial sections in layer 1). To visualize the labeled dendrites in 3D, electron micrographs were aligned consecutively using Photoshop (Adobe) and then the stack of serial images exported to a Silicon Graphics workstation running the DepthAnalyser module in Imaris Software (Bitplane AG, Zurich, Switzerland). Labeled elements were outlined on each serial section and the completed structure exported to 3D Studio Max (Discreet Software, USA) for color rendering. Supplementary Information: Electrophysiology Sharp electrode intracellular recordings were performed on wild type c57/bl6 five week old male mice (n = 15) essentially as described 34. Chessboard deprived mice had the appropriate whiskers trimmed to < 1 mm 3-5 days before the acute experiment. Receptive fields were constructed by measuring sensory stimulation-evoked synaptic potentials (PSPs). For each whisker 25 stimulus trials were collected with an interstimulus interval of 4 seconds. Neurons (14/15) were recovered and assigned to a barrel column using standard histological techniques 34. Recordings from layer 2/3 (12/15) and layer 5 (3/15) neurons were pooled since the time-course and extent of plasticity is similar in these layers 5. Receptive fields were computed using the amplitude of the averaged PSP for each whisker. Individual responses were median filtered to remove action potentials. Since the membrane potential fluctuates between up and down states, where the up state corresponds to massive and synchronous network excitation 23, 34, trials in which the neuron was in an up-state during stimulation were removed. To define up-states, an allvalue histogram was computed for the membrane potential measurements during each stimulus trial. The histogram typically had two peaks corresponding to up and down states. If, during stimulus onset, the membrane potential was within range of the peak at higher potentials the trace was removed from the average. Whisker-evoked PSPs were identified as the largest value within a 50 ms poststimulus window. PSP amplitudes varied greatly between animals (range: 3.3 – 27 mV). Surround 1 and 2 (1SW and 2SW, respectively) PSPs were defined as the responses to the adjacent whiskers one or two rows or columns away from the PW. Supplementary Information: Analysis of spine volumes and spine shapes Analysis of spine volumes was performed on a subset of spines that were well-separated from dendrites (n=301, 6 animals). Relative volumes were estimated as the spine fluorescence divided by the dendritic fluorescence 29. This estimate assumes that GFP is homogeneously disitributed throughout the cytoplasm and that dendritic shafts do not vary much in diameter. Using this estimate we find that spine volumes vary by a factor of > 50, consistent with serial section EM reconstructions where spines in adult hippocampus vary by ~ 100 fold in volume 30, 42. Stable spines were on average 75 % larger than transient spines (p < 0.05) (Supplementary Fig. 2a). Of the spines suitable for volume analysis, stable spines comprised 61% of the population, consistent with the total spine population (Supplementary Fig. 2b). 77% of stable spines had a well-defined spine head, distinct from the spine neck. Conversely, 83% of spines that had a well-defined head were stable, and less than 4% were transient (Supplementary Fig. 2c). We define mushroom spines as spines with a well-defined spine head that had large spine volumes (within 50% of the largest spine volumes measured). We estimate that this corresponds to the larger half of mushroom spines defined using ultrastructural criteria 30. These large mushroom spines were found to be exceptionally stable, with 93% surviving as long as they were imaged (8-32 days; Fig. 5a,d; Supplementary Fig. 2d, e). Relative Volume a b c * d Spines with well-defined heads All spines Large mushroom spines 10 60.5% 5 0 e Spine Volumes 15 1 day 2-7 days 8 days Transient Semi-stable Stable Day 1 Day 2 Day 3 Day 4 1 day Day 5 92.9% 82.9% Day 6 2-7 days Day 7 Day 8 8 days Day 29 Legend Transient Stable Supplementary Figure 2. Larger spines are relatively stable. a, Spine head volumes as a function of spine lifetimes. Stable spines were significantly larger than transient and semi-stable spines (p < 0.05). b, Breakdown of spine lifetimes for the subset of spines selected for volume analysis (n = 301). 61% of spines existed for at least 8 days (stable). c, Breakdown of spine lifetimes for spines with well-defined spine heads (e.g. red arrow and yellow arrowhead in e; n = 170). 83% of spines with well-defined spine heads were stable. d, Breakdown of spine lifetimes for large mushroom spines (yellow arrowhead, but not red arrow; n = 14). All but one mushroom spine existed for as long as imaged. e, Examples of stable mushroom spines (e.g. yellow arrowhead) and small transient spines (e.g. blue arrowheads). Scale bar, 5μm.