TPJ_4363_sm_Legends
advertisement

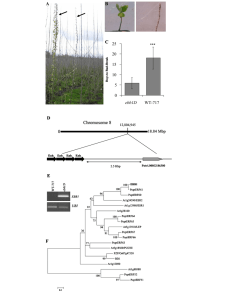
Supplementary figure legends Figure S1: Schematic representation of the position of the PCR amplification fragments used in ChIP-semi-quantitative PCR (ChIP-PCR) or ChIP-quantitative PCR (ChIP-qPCR) analysis relative to the putative transcription start site of the gene (indicated by arrow and +1). Horizontal bars represent the ChIP-qPCR amplified fragments. When the ChIP-qPCR fragment overlaps with ACII DNA-binding sites, then the corresponding motif sequence is indicated. Figure S2: The ZmA1 gene is a direct target of ZmMYB31. qPCR analysis of ChIP DNA enrichment performed with purified ZmMYB31-specific antibody. Enrichment of the ZmA1 promoter region is relative to the enrichment of the Copia gene. Error bars represent the standard error of three biological replicates. The average values are shown, and the error bars indicate the standard deviation of the samples. Statistical analysis of differences between samples was performed using a Student’s t test. *, Significant at p < 0.05. Supplementary Table legends Table S1: SELEX sequences used for ZmMYB31 (26bp): The consensus sequences are shown in red and underlined. Table S2: ZmMYB31 overexpression in A. thaliana induces the synthesis of stress-related proteins. Panel (a) corresponds to downregulated proteins (D1 to D11) in transgenic plants and panel (b) to induced proteins (U1 to U8) in transgenic plants. Theor. pI and Exp. pI refer to theoretical and experimental isoelectric point. Theor. MM and Exp. MM: theoretical and experimental molecular mass. Pept nº: peptide number. Table S3: Sequence of the gene-specific primers used in this work.