Genome architectures of Bartonella grahamii strains isolated from
advertisement

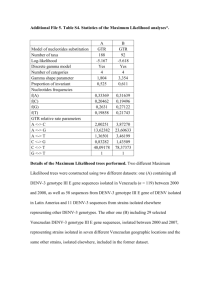
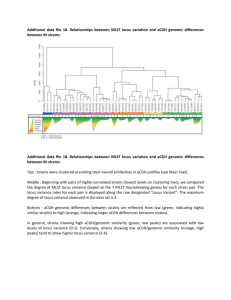
Genome architectures of Bartonella grahamii strains isolated from Swedish rodents Marcel Verhaart Bartonella grahamii belongs to the bacteria genus Bartonella. This genus consists of 16 species including some well-known human pathogens such as B. henselae and B. quintana. B. grahamii itself is normally found in small rodents like mice and a few cases of human infections are known. For my project I used 17 strains originating from three different locations in central Sweden as well as a strain whose entire DNA was sequenced recently, originating from Sigtuna. These strains were isolated from different mouse species. I have estimated the size of the chromosome in these strains. All strains had chromosomes of similar size (containing about 2.3 million base pairs). By cleaving this DNA with enzymes recognizing particular sequences of base pairs (such as the enzyme NotI), I could see that the strains showed cleavage patterns typical for the different locations, not for the host from which they were isolated. Strains isolated from Håtunaholm all showed the same NotI pattern, which was clearly different from the strains from the other locations. The strains isolated from Ålbo and Kumla seemed to be more closely related, although they still showed some differences in their cleavage pattern. The sequenced strain showed almost the same pattern as the strains from Håtunaholm, which is logical because this is the closest location to Sigtuna. This indicated that the evolution within this species is not host specific but location specific. With another technique permitting me to compare DNA sequences in small regions of the chromosome (Southern hybridization), I found some differences in genome content in a region in the genome where a lot of differences between strains have been found in other species. It is likely that more differences extending these experiments to other regions of the chromosome. An interesting finding was that there seemed to be an extra NotI cleavage site in this part of the genome in some of the strains. Degree project in Biology Examensarbete i biologi, 20 p, vår 2005 Department of Biology Education and Department of Molecular Evolution, Uppsala University Supervisor: Siv Andersson