FWB_12038_sm_AppendixS1
advertisement

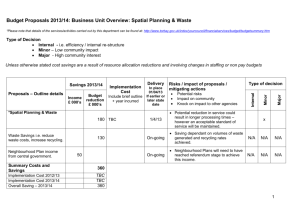
Fuentes-Rodríguez: Macroinvertebrate diversity in farm ponds Appendix S1 Description of the procedure implemented to generate Moran’s eigenvector maps (MEM) spatial variables. MEMs are an extension of the previously developed approach known as principal coordinates of neighbor matrices (PCNM; Borcard & Legendre, 2002; Borcard et al., 2004). Both methods produce a set of orthogonal spatial variables (eigenvectors), that can be used as predictors in multiple regression or redundancy analysis (RDA), but MEM variables are more flexible in case of geographically irregular distribution of sites (Sattler et al., 2010), as it was the case in our ponds. The more general framework defined by MEM differ from PCNM in two ways: (1) different connectivity matrices can be applied to define the links among sampling sites, (2) different spatial weighting functions among sampling sites can be used. Both connectivity and weighting matrices define the final spatial weighting matrix, which may be used as a predictor to model spatial variation of biological data. The choice of both connectivity and weighting matrices is a critical step since it affects the outcome results of the spatial analysis. As suggested by Dray, Legendre & Peres-Neto (2006), we applied a data-driven approach to select the spatial weighting matrix consisting in the following steps: (1) to define a set of possible spatial weighting matrices (obtained from the different combinations of connectivity matrices and weighting functions); (2) to compute MEM for each of these models, (3) to compute RDA of the multivariate biological data with each MEM model and retain those eigenvectors that result in the most appropriate model according to the corrected Akaike Information Criterion (AICc, Burnham & Anderson, 2002) and finally (4) to select the model with the lowest AICc. Five ways of defining neighbor networks were examined (Dray, Legendre & PeresNeto, 2006): Delaunay triangulation (tri), Gabriel graph (gab), relative neighbourhood graph (rel), minimum spanning tree (mst) and distance criterion (dnn). For the last case, Fuentes-Rodríguez: Macroinvertebrate diversity in farm ponds two sites i and j were considered as neighbourghs if dij < k, where dij is the Euclidean distance between sites and k is the threshold distance. Ten values of the parameter k were considered, which were evenly distributed between the shortest value that keeps all sites connected and the highest distance at which an empirical multivariate variogram (Wagner, 2003) is significant (Dray, Legendre & Peres-Neto, 2006). To construct the matrix of weights, we assumed that similarity in assemblages’ composition is higher for pairs of ponds that are spatially closer. Thus, ecological similarity was supposed to decrease with distance according to the function f = 1 – (dij/max(dij))y, where max(dij) is the maximum distance defined within a given neighbour network and y is a parameter. We considered the sequence of integers between 1 (linear decay of similarity with distance) and 10 (different concave-down spatial relationships) for y. We computed MEM for the five types of binary connectivity networks (bin) and for the combinations of these connectivity matrices with the weighting function and identified the most appropriate spatial weighting matrix according to AICc. References Borcard D. & Legendre P. (2002) All-scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecological Modelling, 153, 51-68. Borcard D., Legendre P., Avois-Jacquet C., Tuomisto H. (2004) Dissecting the spatial structure of ecological data at multiple scales. Ecology, 85, 1826-1832. Burnham K.P. & Anderson, D.R. (2002) Model selection and multimodel inference: a practical information-theoretic approach. 2nd Edition Springer-Verlag, New York, USA. 488 p. Fuentes-Rodríguez: Macroinvertebrate diversity in farm ponds Dray S., Legendre P. & Peres-Neto P.R. (2006) Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecological Modelling, 196, 483-493. Sattler T., Borcard D., Arlettaz R., Bontadina F., Legendre P., Obrist M.K. & Moretti M. (2010) Spider, bee, and bird communities in cities are shaped by environmental control and high stochasticity. Ecology, 91, 3343-3353. Wagner H.H. (2003) Spatial covariance in plant communities: integrating ordination, geostatistics, and variance testing. Ecology, 84, 1045-1057.