Structure determination of E2(TG) and alignment with E1Ca2+
advertisement

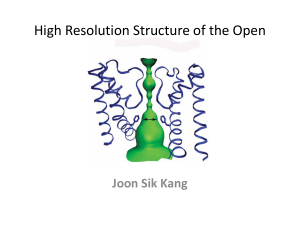
Supplementary Information 1 Structure determination of E2(TG) and alignment with E1Ca2+ Modelling was started by determining the axis of rotation of the N-domain with respect to the P-domain with the program Dyndom16. For this purpose, we used the models for E1Ca2+ (1EUL) and the Mg/F complex (Toyoshima et al., unpublished). (The latter model was made in a similar way to the present model but started from the one for the tubular crystals10 in which the enzyme is in a state similar to the Mg/F complex (i.e. E2P)13, and the axis determination was therefore unnecessary.) If the N and P-domains were used for the search model and the orientation of the N-domain was the same as in the Mg/F complex, no definite solution was found by PC-refinement35. But 10° and 20° rotations of the N-domain around the axis thus determined gave the same and unique solution. The A-domain was then included in the PC-refinement. This increased the correlation coefficient (E2E2 target) substantially (Table 1). Then the NCS-related counterpart was located by calculating combined translation functions using CNS37. At this stage, most of the electron density of a whole molecule, in particular the part near the cytoplasmic domains was well defined after solvent flattening using Solomon36 and was easily interpreted. The electron density map showed that the N and P domains were virtually identical to those in E1Ca2+ (whereas the P-domain in the Mg/F complex showed large conformation changes) and that the A-domain required modifications near the interface with the P-domain. Ten transmembrane helices were evident and the density representing TG was clear at this stage. The models for the M4 and M5 helices were built into the electron density and included first, because these two helices are integrated into the P-domain and therefore well defined. M7-M10 helices were added next; M1-M3 helices contain long loops connecting to the A-domain and were therefore added last., assuring the correctness of the model. The model was gradually expanded by iterating PC refinement, rigid body refinement, map calculation, solvent flattening and manual model building using Turbo-Frodo. The model previously built (i.e. 1EUL) was useful Supplementary Information 2 for map interpretation and reassuring the correctness of the model; the loops were, however, built from scratch because they were often less ambiguous than in E1Ca2+. To check the correctness of the model even further, we tried simple molecular replacement solution of the P21 crystal form that diffracted to 3.3 Å resolution using the entire model from the beginning. After two cycles of simulated annealing refinement using CNS, Rfree went down to 28.8%, confirming the quality of the model. For initial alignment of the two structures, M7-M10 transmembrane helices were used, because their relative positions were essentially the same (rms difference 0.73 Å; see also animations 1 and 2). Then the alignment was refined using all the C atoms within 1 Å distance. This revealed important residues that do not move, such as Glu771 on M5, one of the Ca2+-binding residues. Also, in either structure, the orientations of the M7-M10 helices with respect to the crystallographic a-axis were roughly the same. Therefore the ab-plane of the present P41 crystal was assumed to coincide with the membrane plane. This orientation is also the same as that in the tubular crystal in which we could see the position of the lipid bilayer. In the E2(TG) structure, the position of the cytoplasmic surface of the bilayer is rather well defined, because a part of M1 helix (M1’ in Fig. 2) is now kinked to form an amphipathic helix (D59 is shown in Fig. 1) presumably lying on the bilayer surface. This position is consistent with those of the side-chains of Arg, Lys (R63 on M1 is shown in Fig. 1) and Trp residues (W832 on M7 is shown in Fig. 1). Although exogenous phospholipids were added in crystallisation, no clear electron densities representing lipid molecules were observed. Table 1. Progress of the PC and rigid body refinements model number of correlation packing Supplementary Information 3 atoms in the coefficient model (E2E2)$ P+N 3088 0.161 0.088 +A 4279 0.215 0.121 + NCS mate 8558 0.386 0.241 +M4,M5 9748 0.448 0.275 main part 12868 0.481 0.365 final 15434 0.799 0.435 $Calculated for 15-4 Å resolution. function