Alpha-helical transmembrane protein fold prediction using residue contacts
advertisement
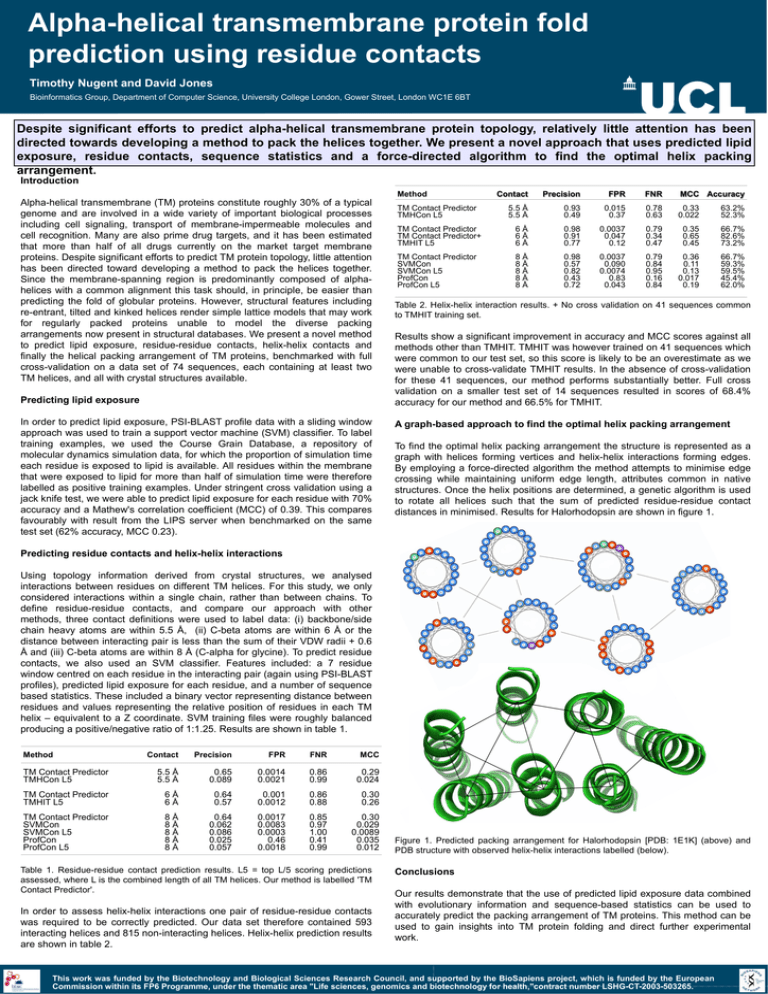
Alpha-helical transmembrane protein fold prediction using residue contacts Timothy Nugent and David Jones Bioinformatics Group, Department of Computer Science, University College London, Gower Street, London WC1E 6BT Despite significant efforts to predict alpha-helical transmembrane protein topology, relatively little attention has been directed towards developing a method to pack the helices together. We present a novel approach that uses predicted lipid exposure, residue contacts, sequence statistics and a force-directed algorithm to find the optimal helix packing arrangement. Introduction Method Alpha-helical transmembrane (TM) proteins constitute roughly 30% of a typical genome and are involved in a wide variety of important biological processes including cell signaling, transport of membrane-impermeable molecules and cell recognition. Many are also prime drug targets, and it has been estimated that more than half of all drugs currently on the market target membrane proteins. Despite significant efforts to predict TM protein topology, little attention has been directed toward developing a method to pack the helices together. Since the membrane-spanning region is predominantly composed of alphahelices with a common alignment this task should, in principle, be easier than predicting the fold of globular proteins. However, structural features including re-entrant, tilted and kinked helices render simple lattice models that may work for regularly packed proteins unable to model the diverse packing arrangements now present in structural databases. We present a novel method to predict lipid exposure, residue-residue contacts, helix-helix contacts and finally the helical packing arrangement of TM proteins, benchmarked with full cross-validation on a data set of 74 sequences, each containing at least two TM helices, and all with crystal structures available. Predicting lipid exposure In order to predict lipid exposure, PSI-BLAST profile data with a sliding window approach was used to train a support vector machine (SVM) classifier. To label training examples, we used the Course Grain Database, a repository of molecular dynamics simulation data, for which the proportion of simulation time each residue is exposed to lipid is available. All residues within the membrane that were exposed to lipid for more than half of simulation time were therefore labelled as positive training examples. Under stringent cross validation using a jack knife test, we were able to predict lipid exposure for each residue with 70% accuracy and a Mathew's correlation coefficient (MCC) of 0.39. This compares favourably with result from the LIPS server when benchmarked on the same test set (62% accuracy, MCC 0.23). Contact Precision FPR FNR MCC Accuracy 5.5 Å 5.5 Å 0.93 0.49 0.015 0.37 0.78 0.63 0.33 0.022 63.2% 52.3% TM Contact Predictor TM Contact Predictor+ TMHIT L5 6Å 6Å 6Å 0.98 0.91 0.77 0.0037 0.047 0.12 0.79 0.34 0.47 0.35 0.65 0.45 66.7% 82.6% 73.2% TM Contact Predictor SVMCon SVMCon L5 ProfCon ProfCon L5 8Å 8Å 8Å 8Å 8Å 0.98 0.57 0.82 0.43 0.72 0.0037 0.090 0.0074 0.83 0.043 0.79 0.84 0.95 0.16 0.84 0.36 0.11 0.13 0.017 0.19 66.7% 59.3% 59.5% 45.4% 62.0% TM Contact Predictor TMHCon L5 Table 2. Helix-helix interaction results. + No cross validation on 41 sequences common to TMHIT training set. Results show a significant improvement in accuracy and MCC scores against all methods other than TMHIT. TMHIT was however trained on 41 sequences which were common to our test set, so this score is likely to be an overestimate as we were unable to cross-validate TMHIT results. In the absence of cross-validation for these 41 sequences, our method performs substantially better. Full cross validation on a smaller test set of 14 sequences resulted in scores of 68.4% accuracy for our method and 66.5% for TMHIT. A graph-based approach to find the optimal helix packing arrangement To find the optimal helix packing arrangement the structure is represented as a graph with helices forming vertices and helix-helix interactions forming edges. By employing a force-directed algorithm the method attempts to minimise edge crossing while maintaining uniform edge length, attributes common in native structures. Once the helix positions are determined, a genetic algorithm is used to rotate all helices such that the sum of predicted residue-residue contact distances in minimised. Results for Halorhodopsin are shown in figure 1. Predicting residue contacts and helix-helix interactions Using topology information derived from crystal structures, we analysed interactions between residues on different TM helices. For this study, we only considered interactions within a single chain, rather than between chains. To define residue-residue contacts, and compare our approach with other methods, three contact definitions were used to label data: (i) backbone/side chain heavy atoms are within 5.5 Å, (ii) C-beta atoms are within 6 Å or the distance between interacting pair is less than the sum of their VDW radii + 0.6 Å and (iii) C-beta atoms are within 8 Å (C-alpha for glycine). To predict residue contacts, we also used an SVM classifier. Features included: a 7 residue window centred on each residue in the interacting pair (again using PSI-BLAST profiles), predicted lipid exposure for each residue, and a number of sequence based statistics. These included a binary vector representing distance between residues and values representing the relative position of residues in each TM helix – equivalent to a Z coordinate. SVM training files were roughly balanced producing a positive/negative ratio of 1:1.25. Results are shown in table 1. Method Contact Precision FPR FNR MCC TM Contact Predictor TMHCon L5 5.5 Å 5.5 Å 0.65 0.089 0.0014 0.0021 0.86 0.99 0.29 0.024 TM Contact Predictor TMHIT L5 6Å 6Å 0.64 0.57 0.001 0.0012 0.86 0.88 0.30 0.26 TM Contact Predictor SVMCon SVMCon L5 ProfCon ProfCon L5 8Å 8Å 8Å 8Å 8Å 0.64 0.062 0.086 0.025 0.057 0.0017 0.0083 0.0003 0.46 0.0018 0.85 0.97 1.00 0.41 0.99 0.30 0.029 0.0089 0.035 0.012 Table 1. Residue-residue contact prediction results. L5 = top L/5 scoring predictions assessed, where L is the combined length of all TM helices. Our method is labelled 'TM Contact Predictor'. In order to assess helix-helix interactions one pair of residue-residue contacts was required to be correctly predicted. Our data set therefore contained 593 interacting helices and 815 non-interacting helices. Helix-helix prediction results are shown in table 2. Figure 1. Predicted packing arrangement for Halorhodopsin [PDB: 1E1K] (above) and PDB structure with observed helix-helix interactions labelled (below). Conclusions Our results demonstrate that the use of predicted lipid exposure data combined with evolutionary information and sequence-based statistics can be used to accurately predict the packing arrangement of TM proteins. This method can be used to gain insights into TM protein folding and direct further experimental work. This work was funded by the Biotechnology and Biological Sciences Research Council, and supported by the BioSapiens project, which is funded by the European Commission within its FP6 Programme, under the thematic area "Life sciences, genomics and biotechnology for health,"contract number LSHG-CT-2003-503265.