Molecular factor VIII gene identification and Hemophilia A in Lebanon
advertisement

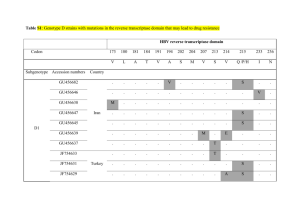
Administrative Information المعلومات االدارية :المرجع Project Title - )عنوان المشروع (عربي وأجنبي Molecular factor VIII gene identification and Hemophilia A in Lebanon: Genotype-phenotype correlation. Which relation between molecular anomalies and risk of inhibitor? : يف لبنانA ) لداء الناعور ( هيموفيلياVIII ملورثة (جينة ) العامل ّ تعني شذوذ اجلزيئيات ما هي العالقات بني شذوذ اجلزيئيات وخطر الكابثات (املتبطّات ) ؟. عالقة نـمط وراثي – نـمط ظاهري Principal Investigator - الباحث الرئيسي رقم الهاتف العنوان االلكتروني Telephone e-mail 03704864 claudiakhayat@yahoo.fr العنوان Address الوظيفية Post Enseignant المؤسسة Institution Universite Saint Joseph االسم Name Claudia DJAMBAS KHAYAT Co-Workers - الباحثون المشاركون العنوان االلكتروني e-mail 2005-2007 المؤسسة Institution االسم Name : Duration -المدة التعاقدية للمشروع Scientific Information العلمية المعلومات ّ Objectives - الهدف Our objective was to identify the spectrum of mutations of the F8 gene responsible of the HA in Lebanese patients, and perform a genotype/phenotype correlation in the patients’ cohort. Achievements -أالنجازات المحققة A cohort of 76 Hemophilia A (HA) patients from 52 unrelated families was studied, representing almost 87% of the HA registrated patients in Lebanon. We were able to identify all causative mutations. Twenty three mutations identified in this study were novel: 12 missense, 3 nonsense, 2 splice site, 5 small deletions and one large deletion. A history of inhibitor found in 7 over 75 patients correlated with large deletion, intron 22 inversion, and nonsense mutations. Perspectives - آفاق البحث The knowledge of causative mutation of our HA patient is very important, particularly because it represents a huge step for genetic counselling. Indeed 82% of our cases were familial and hemophilia care has poor resources in our country. The characteristics of the mutation responsible for the disease, being the strongest predictor of the risk of inhibitor development so far identified, will also light up our modalities treatments and could help to minimize the risk of this complication and improve treatment outcomes. Publications & Communications - المنشورات والمساهمات في المؤتمرات 1- Djambas Khayat, N. Salem, E. Chouery, S. Corbani , I. Moix, E. Nicolas, M. A. Morris, P. De Moerloose , A. Megarbane. . Detection of 23 novel mutation in Lebanese hemophilia population. Poster, XXI International Society on Thrombosis and Haemostasis congress, July 2007 Geneva, Suitzerland 2- C. Djambas Khayat, N. Salem, E. Chouery, S. Corbani, I. Moix, E. Nicolas, M. A. Morris, P. De Moerloose , A. Megarbane. Molecular Analysis of F8 in Lebanese Hemophilia A Patients: Novel Mutations and Phenotype-Genotype Correlation . Oral presentation, 2nd Pan Arab Human genetic Conference. November 2007, Dubai United Arab Emirat. 3- Djambas Khayat C, Salem N, Chouery E, Corbani S, Moix I, Nicolas E, Morris MA, DE Moerloose P, Mégarbané A. Molecular analysis of F8 in Lebanese haemophilia A patients: novel mutations and phenotype-genotype correlation. Article: Haemophilia. 2008 May 12. 12. Abstract - موجز عن نتائج البحث Summary. Haemophilia A (HA) is an X-linked recessive hereditary bleeding disorder affecting one in 5000 men, resulting from mutations in the F8 gene. Our objective was to identify the spectrum of mutations of the F8 gene in Lebanese patients, and to perform genotype/phenotype correlations. A group of 79 HA patients from 55 unrelated families was studied. Patients were screened for intron 22 and intron 1 inversion using PCR. In the absence of mutations in both introns, a dHPLC screening followed by a DNA sequencing of all coding regions was performed. When patients presented novel mutations, 150 control chromosomes were tested to exclude common polymorphisms. Large deletions were confirmed by MLPA technique. The mRNA was specifically studied whenever a splice site mutation was detected. In addition, studies of the putative biochemical function and FVIII 3D structures were conducted. Thirty-four mutations were identified in this study of which 21 were novel: 11 missense, two nonsense, two splice sites, five small deletions and one large deletion. Inhibitor found in three over 75 patients correlated with large deletion, intron 22 inversion, and nonsense mutations. We were able to identify all causative mutations in those HA patients. This knowledge represents a huge step for genetic counselling.