heredity and gene action/chromosomes and gene function
advertisement

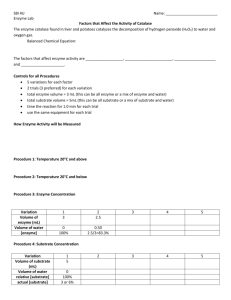
BASIC BIOCHEMISTRY/MOLECULES OF LIFE THE ENZYMOLOGY OF β-GALACTOSIDASE II CALCULATING: Km and Vmax for β-galactosidase In practical number 2 we studied the effect of pH, which affects the 3-D structure of the protein and the chemistry of the active site (and substrate), on the enzyme β-galactosidase. This enzyme normally catalyses the breakdown of lactose in the lactose metabolic pathway, but the catalytic activity of the enzyme can be monitored by following the hydrolysis of a colourless artificial substrate ONPG to galactose and orthonitrophenol because the latter is a yellow colour and can be monitored using a spectrophotometer set at 420 nanometers. (Please see below). Not only is a protein’s function inextricably linked to the pH of the surrounding medium, but the rate of catalysis is also dependant upon the interaction of the enzyme and its substrate. The study of enzyme reaction rates is referred to as enzyme kinetics and today we are going to meet the two most fundamental parameters used in the description of enzyme kinetics: The enzyme’s rate constant (Km) and its speed of reaction (Vmax). These were first explored by Michaelis and Menten. They are therefore referred to as: the Michaelis-Menten constants. See http://oregonstate.edu/instruction/bb450/lecturenoteskevin/highlightsenzymes.html Enzyme activity can generally be described by the following formula: Where: E = Enzyme. S = Substrate. ES = Enzyne-substrate complex. P = product. Within this context: The rate of formation of ES = k1[E][S] The rate of breakdown of ES = k-1[ES] (backwards reaction) + k2[ES] (forward reaction) Vmax: This is simply the maximum possible speed of the enzyme’s reaction. It is experimentally ascertained by increasing the substrate concentration until the rate of catalysis reaches its maximum speed. Vmax can therefore be experimentally estimated by measuring the rate of reaction as substrate levels are increased. Km: Steady state conditions refer to conditions under which the concentration of ES is relatively constant. Under these circumstances, the rates of formation of ES, and of breakdown of ES, are equal. In other words: k1[E][S] = k-1[ES] + k2[ES] The rate constants (k1, k-1, k2) do not change for any given enzyme and so a simple mathematical trick can be used to combine them into one composite constant referred to as Km. Km is defined as: (k-1 + k2)/k1) Not surprisingly different enzymes react in very different ways with their substrates and so Km values vary significantly from one enzyme to another. It is therefore a very useful tool for exploring the mechanism by which an enzyme carries out its catalytic activity. Estimating Vmax and Km: As you will have heard in your lectures Vmax and Km are intimately related. Vmax can be experimentally determined allowing Km to be estimated. One way of doing this is by using a double reciprocal plot of substrate concentration and reaction velocity, referred to as the Lineweaver-Burk plot. Reciprocals of experimentally determined reaction velocities (1/v) for a range of substrate concentrations are plotted against the reciprocal of substrate concentration (1/[S]). The y-intercept provides the reciprocal of Vmax, from which the Vmax can be determined. The slope can be calculated and this is equal to Km divided by Vmax. Given that Vmax is known – Km can then be calculated. For an interactive version of such a plot: http://cti.itc.virginia.edu/~cmg/Demo/kinetics/lb/lb/lb.html Here we utilize the ONPG-based absorbance based assay for β-galactosidase to determine experimentally the Vmax and Km for this enzyme. CHARACTERISING ENZYME ACTIVITY BY VARYING SUBSTRATE CONCENTRATION. 1A. ONE HOUR PREPARATORY SESSION. Objectives: To re-enforce the use of a spectrophotometer and the accurate use of micropipettes. To explore Km and Vmax. To plan a series of reaction conditions to test the effect of substrate concentration on the enzyme activity of β-galactosidase. To become familiar with how to use a Lineweaver-Burk plot to calculate Vmax and Km. Resources: A spectrophotometer, a P1000 micropipette and tips. This handout. Information on a number of solutions. Your group members and a facilitator. PROCEDURE: 1. Ensure that you are familiar with the correct use of the spectrophotometer allocated to your bench and the P1000 micropipette. 2. Use the information presented in the introduction and in the paragraph below to explore the relationship between the speed of an enzyme reaction and the concentration of substrate available to it. Record this in your lab book. Questions: When an enzyme has a high affinity for its substrate will the rate of ES formation be high or low? Will the enzyme’s Km be high or low? Why? Will Km vary if the concentration of enzyme is halved? Why? Record this in your practical book. Just as the affinity of an enzyme for its substrate affects the speed of product accumulation so too the level (and substrate levels can fluctuate widely in biological systems) of substrate availability also affects product accumulation. Consider product accumulation if there is no substrate present. Now consider the speed of product accumulation as the level of substrate increases from very low levels to extremely high ones. Sketch a graph of rate of product accumulation versus concentration of substrate. What eventually limits the velocity of product accumulation? Can you suggest why this maximum velocity cannot be increased by increasing the concentration of substrate? Will the maximum velocity vary if the concentration of enzyme is halved? Why? Record this in your practical book. These two parameters (Km and Vmax) are intimately related to one another. Draw in this relationship in on your sketched graph. We can ascertain the affinity of an enzyme for its substrate by establishing the Vmax of an enzyme in the laboratory. Simply alter the concentration of substrate in an enzyme reaction and accurately measure the initial speed (in this practical within 30 seconds) of product accumulation until the further addition of substrate does not increase the speed of product accumulation. Why is it important to use early estimations of rate of product accumulation? (Consider how decreasing levels of substrate might affect the accumulation of ES). In practical 3 we shall focus our attention on how altering the substrate concentration affects the velocity of this enzyme. 3. Use the following information to draw up a table for the preparation of solutions containing different concentrations of ONPG. The molecular mass of ONPG is 352. 70.4 mgs were weighed out and dissolved in 20ml of water. This is the stock solution of ONPG. Find out the molarity of ONPG in this solution and use that and the table below to plan how to prepare 5ml solutions containing 5mM, 2.5mM, 1.5mM, 1.0mM, 0.5mM ONPG concentrations respectively. Draw up this dilution table of ONPG stock and water in your book: Final Concentration of 5mM ONPG ONPG required in your 5ml stock solutions. Dilution factor required (ie Stock ONPG must be diluted by…). Final volume required. 5ml Volume of stock ONPG solution required. Volume of water required. 2.5mM ONPG 1.5mM ONPG 1.0 mM ONPG 0.5mM ONPG 5ml 5ml 5ml 5ml 4. Prepare a table of reaction mixtures using the following information. In the practical exercise you will be supplied with: 0.48M Tris Acetate 0.48M phosphate buffer pH 7.5. 1.2 units/ml of β-galactosidase (And you will prepare the 5 ml solutions of ONPG.) Given that you will add of 0.5ml of the appropriate ONPG solution to each 3ml reaction tube. Prepare a table of 3ml reaction volumes such that the final concentrations of the other constituents are as follows: 0.08M Tris Acetate 0.08M phosphate buffers at pH 7.5. 0.2 units/ml of β-galactosidase. Sample 1 5mM ONPG Sample 2 2.5mM ONPG Sample 3 1.5mM ONPG Sample 4 1.0mM ONPG Sample 5 0.5mM ONPG 0.5ml 0.5ml 0.5ml 0.5ml 0.5ml 3ml 3ml 3ml 3ml 3ml Volume of 0.48M Tris Acetate Volume of 0.48M phosphate buffer Volume of appropriate ONPG solution. 1.2 units/ml of βgalactosidase H2O Total Volume You will also need to set up two controls to check that the substrate and enzyme are BOTH required for the reaction to occur. What will they be? Add these to your book. Sample 6 Control (substrate) Sample 7 Control (enzyme) Volume of 0.48M Tris Acetate Volume and pH of 0.48M phosphate buffer 5mM ONPG 2.4 units/ml of β-galactosidase Total Volume 5. Familiarise yourself with how to obtain reciprocal values in order to prepare for plotting the graph and how to obtain the slope of the line. 1B. 90 MINUTE INDIVIDUAL SESSION Objectives: To gain experience in the use of a spectrophotometer and in the accurate use of micropipettes. To explore the effect of substrate concentration on enzyme activity. To use this information to calculate the Km and Vmax for β-galactosidase. Resources: Your laboratory notebook – updated with information from the one hour group preparation session. A spectrophotometer set at 420nM and 3ml cuvettes. P1000 micropipettes and tips. A solution of 1.2units/ml of β-galactosidase. A stock solution of ONPG. Universal tubes and prepared solutions. A facilitator. PROJECT: Use the table in your notebook to set up appropriate dilutions of ONPG using distilled water and the ONPG solution supplied. Set up 5 cuvettes containing: the acetate, Tris buffer, and ONPG at five different concentrations. Label these 1 to 5 and leave on your bench. Zero your spectrophotometer by inserting an appropriate control tube (see last experiment) and pressing the DARK BLUE button. NB. ALWAYS ENSURE THAT THE CUVETTES HAVE BEEN DRIED AND ARE FACING THE CORRECT WAY SO THAT THE CLEAR FACES ARE IN THE LIGHT PATH WHEN USING SPECTROPHOTOMETERS. Now you are ready to proceed with the five samples. Add the appropriate volume of enzyme to tube number 1 mix and replace the blank with it. Press the GREEN button. Take the readings (to the nearest 0.1) every 30 seconds for two minutes. Record these in a table in your book without delay Zero the machine again. Repeat for samples 2-4 and your controls. Use the data from each sample to plot five graphs of activity as increase in absorbance at 420nM (O/D 420) against time (secs). Use the initial velocity (the straight line section of each graph) to work out the rate of ONPG hydrolysis in the first 30 seconds. Convert this to rate per minute. Record the concentration of ONPG and its appropriate rate of ONPG hydrolysis per minute) and the rates (if any) of the control samples. Use a calculator to obtain the reciprocal values of the substrate concentrations and the rates. Plot a Lineweaver-Burk plot for these values. Use this straight-line relationship to estimate the Km and Vmax of this enzyme. Pour the samples down the sink and carefully wash out the cuvettes using the plastic wash bottle. Dry the outside carefully and invert on tissue to drain. Wash your hands before leaving the laboratory.