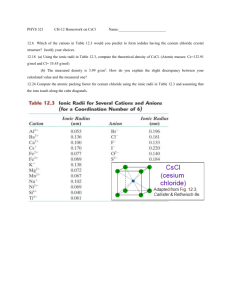
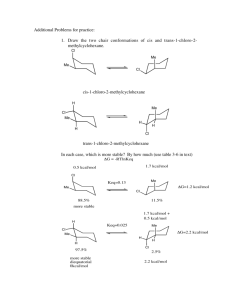
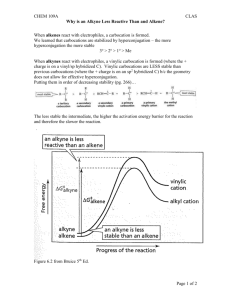
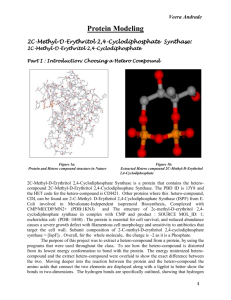
Copyright © 2003 Dr. Ray Luo, All rights reserved.
Unless otherwise indicated, all materials on these pages are copyrighted by Dr. Ray Luo. All rights
reserved. No part of these pages, either text or image may be used for any purpose other than personal use.
Therefore, reproduction, modification, storage in a retrieval system or retransmission, in any form or by
any means, electronic, mechanical or otherwise, for reasons other than personal use, is strictly prohibited
without prior written permission.
BS123A/MB223 Introduction to Computational Biology
Sample Exam Problems
Fall, 2003
Note: This is a close-book/close-note mid-term. You may use a scientific calculator for
any of the problems. Write your solutions on the white papers provided. Remember to
write your name on all papers, and NUMBER all papers.
I. Direction Set Minimization Method
Given a 2-dimensional function f(x, y) = x2 + 3y2 and an arbitrary starting point (1, 2).
Choose the first 1-dimensional line minimization direction to be along i, i.e. the unit
vector direction of x axis.
(a) What is the 1-dimensional function to be minimized?
(b) Compute the first and second derivative of this 1-dimensional function.
(c) Locate the line minimum based on the information in (b), why it is a minimum?
(d) Compute the gradient direction at the minimum, what is the relation of the gradient
direction with the first line minimization direction? I.e. are they parallel, or
perpendicular? Why?
(e) Given the first line minimum from (c), and the direction from (d), perform the second
line minimization following the steps in (b) and (c).
(f) What is the condition for two directions to be conjugate with each other? Are the
directions from second line minimization and first line minimization conjugate with each
other at the first minimum located in (c)?
II. Free Energy Perturbation
(a) In lecture 7, a recipe is given to compute the population ratio of two systems:
N1/N2 = <exp(-(E1-E2)/kBT)>,
where the averaging is over all possible configurations of the two systems. To understand
the numerical limitation of this recipe, consider a case when E1 = -9.1 kcal/mol, and E2 =
-8.8 kcal/mol. Compute the factor exp(-(E1-E2)/kBT) given kBT = 0.6 kcal/mol. Now
consider the second case with E1 = -10.1 kcal/mol and E2 = -1.0 kcal/mol, compute the
factor again.
(b) Finally, suppose we are computing this factor on a computer with limited accuracy,
the computer can only compute a number with accuracy up to the 5th digit after the
decimal point. This means that the absolute error in the computed number is 10-5. What
will be the relative errors of the factor for the above two cases? Which factor can be
computed with relative high confidence by the computer with limited accuracy?
(c) For a hypothetical binding reaction, P + L = PL, P stands for a protein, and L stands
for a ligand. Suppose we know the Gibbs free energies of P, L and PL are GP, GL and
GPL, respectively, how to compute the binding free energy, ΔG?
(d) Suppose free energy perturbation calculation was used to compute the binding free
energy. To make sure that the calculation was reliable, we computed the free energy
changes in two directions. In the direction from P + L → PL, we obtained following data
(in kcal/mol), 1.4, -2.5, -3.0, and -2.1, for the four perturbation steps. In the direction
from PL → P + L, we obtained the data (in kcal/mol), 2.0, 3.1, 2.3, and -1.2 for the four
steps. What are the final binding free energies for the reaction P + L = PL computed in
the above two directions? Give an error estimation in the binding free energy by the
above two calculations.