03. Cells II - D`Youville College

D’YOUVILLE COLLEGE
BIOLOGY 659 - INTERMEDIATE PHYSIOLOGY I
CELL BIOLOGY II
Lecture 3: Nucleus, nucleolus, cellular genetics (chapters 2 & 3)
1. Nucleus:
• chromatin – nucleus is repository of hereditary material (genes), represented by nucleic acid (DNA) combined with protein (= chromatin, nucleoprotein)
• nucleolus – highly staining region of chromatin that makes ribosomal RNA
(some of which is stored in nucleolus)
• nuclear envelope (fig. 2 – 9 & ppt. 1) – double layer of membrane, punctuated by highly structured nuclear pores that control traffic between nucleus & cytoplasm
• molecular biology paradigm – DNA –> RNA –> proteins (fig. 3 – 1 & ppt. 2)
- genes (consisting of DNA) control virtually all activities and characteristics of cell; many genes achieve this by controlling formation of RNAs, which control formation of proteins
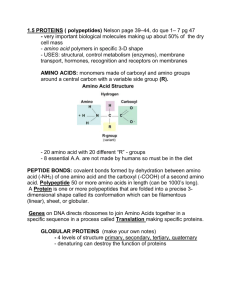
2. Nucleic Acids:
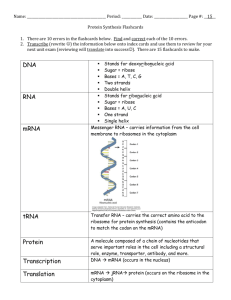
• types: DNA, three kinds of RNA (messenger – mRNA, transfer – tRNA & ribosomal – rRNA); all nucleic acids are polymers of nucleotides
• nucleotides – consist of three components: five-C sugar (deoxyribose in
DNA, ribose in RNA), phosphate & a nitrogenous base (figs. 3 - 3, 3 – 4 & ppts. 3 & 4)
• nitrogenous bases – five different types participate in nucleic acid structure:
2 are 9-membered rings (purines) – adenine & guanine, 3 are 6-membered rings
(pyrimidines) – cytosine, thymine (only in DNA) & uracil (only in RNA)
• complementary pairing – interactions between bases, which H-bond with each other in specific ways: adenine pairs with thymine in DNA (or with uracil in
RNA); cytosine pairs with guanine (fig. 3 – 6 & ppt. 5)
Bio 659 - p. 2 -
• double helix of DNA – nucleotides forming a polynucleotide (nucleic acid) are joined through their phosphate groups (phosphodiester linkage from 5’ carbon of one nucleotide sugar to 3’ carbon of the next); bases on one strand pair with their complementary base in a second nucleotide strand (A with T, C with G) constituting a double-stranded structure, twisted into a double helix (figs. 3 – 2, 3 – 6 & ppt. 6)
3. Replication and Cell Division:
• template copy mechanism – formation of new DNA is believed to involve opening of double-stranded molecule to expose a sequence of bases, which direct assembly of a new second strand through complementary pairing with new nucleotides (replication = DNA synthesis) (ppt. 7)
• DNA polymerase ‘copies’ base sequence of single strands; DNA double helix opens up in several places (replication ‘bubbles’) exposing bases, which then attract nucleotides bearing complementary bases; DNA ligase unites short nucleotide strips from neighboring ‘bubbles’ to produce a complementary strand identical to the original (replica); occurs with both parent strands to produce two identical DNAs each consisting of one new and one parent strand (ppt. 7)
• DNA repair enzymes ‘proofread’ base sequence of new strands & replace erroneous nucleotides with correct ones; occasional mistakes in process can lead to
point mutations – incorrect nucleotides in the sequence, altering the code
• chromosomes – replicated DNA is combined with proteins (nucleoproteins) to make up the chromatin in the nucleus; chromatin becomes coiled & supercoiled, thickening to form chromosomes in preparation for cell division (ppts. 8 & 9)
• mitosis – dividing cells progress through a cell cycle (ppt. 10), when the cell prepares for cell division, DNA replication occurs in cell cycle prior to nuclear division
(mitosis), followed by appearance of double-stranded chromosomes; an identical set of chromosomes is distributed to each daughter cell by a highly orchestrated process called mitosis; stages include prophase, metaphase, anaphase & telophase (fig. 3 – 14 & ppt. 11)
Bio 659 - p. 3 -
4.
Transcription and RNA Functions
:
• genetic code – sequence of bases in nucleotides of DNA represents coded message that specifies pattern of amino acids making up proteins of the cell; each triplet nucleotide sequence represents code word for a particular amino acid (61); three triplets specify ‘stop’ messages, identifying end of gene (table 3 – 1 & ppt. 12)
• synthesis of RNA (transcription) – follows template copy mechanism, like
DNA replication, except isolated strips of only one strand of DNA are copied by the enzyme RNA polymerase (fig. 3 – 7 & ppts. 13 & 14)
• mRNA – working copy of gene; carries sequence of triplets (codons) from each gene to cytoplasmic site to specify sequential assembly of amino acids in protein synthesis (fig. 3 – 8 & ppt. 15); in eukaryotes, introns & exons are present in primary transcript; introns must be removed by ‘cut-and-splice’ process to produce functional mRNA (ppt. 16)
• rRNA – formed in nucleolus of nucleus, combined with proteins in
ribosome formation
• tRNA – cloverleaf-shaped molecules bearing amino acid binding site
(specific for one amino acid) and a loop with a triplet complementary to the codon for its amino acid (anticodon); anticodons of tRNA pair with codons of mRNA at the ribosome (fig. 3 – 9 & ppts. 17 & 18)
Bio 659 - p. 4 -
5.
Translation
:
• proteins – polymers of amino acids; 20 different amino acids may be found in proteins; specific sequence and number of amino acids determines properties of the protein; an immense variety of proteins is possible, each with a unique amino acid sequence
• ribosome – ribonucleoprotein particle, with enzyme activity, bearing binding sites for mRNA & for tRNAs (A site & P site)
• activation of amino acids – formation of amino acyl tRNAs, each with own specific amino acid (fig. 3 - 12 & ppt. 19)
• translation – amino acid sequence is assembled according to genetic code through interactions of mRNA, ribosome & amino acyl tRNAs; special initiation step +
repeating chain elongation steps + termination step (figs. 3 – 9,. 3 – 11 & ppts. 20 & 21)