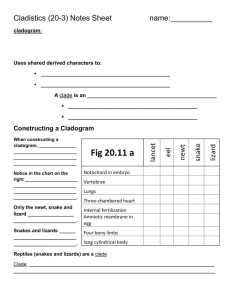
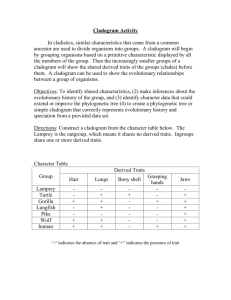
Introduction to cladistics
advertisement

Fossils & Evolution Spring 2012 Due: Tuesday, Feb. 7 Lab Assignment (20 points): Cladistics Introduction to cladistics Cladistics is a method of hypothesizing genealogic relationships among organisms. Like other methods, it has its own set of assumptions, procedures, and limitations. Cladistics is now the most widely used method for phylogenetic analysis and classification, because it provides an explicit and testable hypothesis of evolutionary relationships. The basic idea behind cladistics is that members of a natural group share a common ancestor and are "closely related," more so to one another than to members of other groups. These natural groups, or clades, are recognized by sharing “derived” features that are not present in distant ancestors or other groups. There are two basic assumptions in cladistics: 1. Organisms of a clade are related by descent from a common ancestor. 2. Change in characteristics occurs in lineages over time. The first assumption is a general assumption in evolutionary biology. It essentially means that life arose on earth only once, and therefore all organisms are related in some way. Because of this, we can take any collection of organisms and determine a meaningful pattern of relationships, provided we have the right kind of information. The second assumption, that characteristics of organisms change over time, is the most important assumption in cladistics. It is only when characteristics change that we are able to recognize different lineages or groups. The convention is to call the "original" state of the characteristic “primitive” and the “changed” state “derived.” Cladistic methodology 1. Choose the taxa whose evolutionary relationships interest you. 2. Choose an “outgroup” taxon: i.e., a taxon that does not belong to the clade you are investigating (one that does not possess derived traits shared by members of the clade you are investigating). 3. Determine the characters to be included in the analysis and examine each taxon to determine the character-states for each character. All taxa must be unique. 4. Group taxa by shared derived characteristics, not original or "primitive" characteristics. 5. Work out conflicts that arise by some clearly stated method, usually parsimony. 6. Build a cladogram (tree) following these rules: All taxa go on the endpoints of the cladogram. 1 All cladogram nodes must have one or more derived characteristics that are present in all taxa above the node (unless the character is later modified). All derived characteristics appear on the cladogram only once unless the character state originated more than once by convergent evolution. Parsimony In cladistics, the most parsimonious classification is the simplest one and the one judged to be best supported by data. The simplest classification of a group of taxa is the one that requires the fewest number of transformations from “primitive” character-states to “derived” character-states. For example, consider the following two cladograms (Figure 1). In Cladogram A, amphibians and reptiles are more closely related to each other than either is to fish because both possess limbs, a “derived” trait that evolved in the ancestor of amphibians and reptiles. Cladogram B shows an alternative hypothesis in which fish and amphibians are more closely related to one another than either is to reptiles. According to Cladogram B, however, the “derived” trait limbs evolved twice, once in the ancestor to amphibians and a second time in the ancestor to reptiles. Cladogram A is the more parsimonious hypothesis because it involves only a single origin of the “derived” trait limbs. fish amphibians reptiles fish amphibians reptiles origin of limbs A B Figure 1.—Cladograms showing two alternative hypotheses for relationships among fish, amphibians and reptiles. In PAST software, the acronym “MPT” stands for “most parsimonious tree.” The most parsimonious tree is the one whose length is shortest. Tree length is calculated as the total number of character-state transformations (steps) required to produce a given arrangement of taxa in a cladogram. In the example above, Cladogram A has a shorter tree length than Cladogram B. When dealing with large data sets, multiple MPTs are possible. Assignment—Part 1 (Cladistic analysis of chocolate bars) 1. Begin by working in groups of four. Each group will receive a collection of five “species” of chocolate bars. Construct a data matrix for cladistic analysis by determining character-states for each “species” for the following characters (fill in the following table): Color: 0 = white; 1 = dark 2 Caramel: 0 = absent; 1 = present Nougat: 0 = absent; 1 = present Peanuts: 0 = absent; 1 = present Almonds: 0 = absent; 1 = present DATA M ATRIX color caramel nougat peanuts almonds Hershey’s White (outgroup) Hershey Bar Hershey’s Almond Snickers Snickers Almond 2. Work in pairs once the above table has been filled in. Create and save a file in PAST that contains the same information as the table you’ve just filled in. [Rename rows and columns. Note that PAST does not accept spaces, so use abbreviations.] 3. Create a cladogram. Select all data by clicking the gray button in the upper left corner of the data matrix. Choose “Parsimony analysis” under the Cladistics menu. Use the “Exhaustive” algorithm; “Fitch” optimization; 5 reorderings; 0 Bootstrap replicates). Click the “Go” button to produce the cladogram. [Exhaustive search means that the program will examine all possible trees in order to find the shortest. This is okay for small data sets, but it is impractical for more than about 6 species. There are over 600,000,000 possible trees for a data set of 12 taxa. Fitch optimization means that the program makes no assumptions about character-state transformations: i.e., a character can change from 0 to 1 just as easily as from 0 to 2 (order is unimportant); and a character can change from 0 to 1 just as easily as from 1 to 0 (reversals are acceptable).] How many MPTs did the program find? _______________________ What is the Tree Length of these MPTs? _______________________ Notice that the “Parsimony analysis” window tells you how many trees were evaluated. The “Cladogram lengths” window contains a histogram showing the number of trees of various lengths (e.g., 10 trees that were each 11 steps; 4 trees that were each 10 steps, etc.). How many trees were evaluated __________? 4. Print and label copies of the histogram and MPTs to examine and turn in to the instructor. [It may be necessary to copy and paste into another application such as MS Word or MS PowerPoint. Label each as “Tree 1,” “Tree 2,” etc., as necessary.] 3 5. Examine each of the MPTs. On the trees, label just below the nodes to indicate the character-state transformations at each. The sum of character-state transformations should equal the Tree Length. 6. Are there any instances of convergence (discuss briefly)? 7. Which of the trees do you favor personally (discuss briefly)? Assignment—Part 2 (Cladistic analysis of living and extinct vertebrates) 0 1 1 1 1 1 1 1 1 0 1 1 0 1 1 1 1 1 0 1 1 0 0 1 1 1 1 4 0 0 1 0 0 1 1 0 0 0 0 1 0 0 0 1 0 0 0 0 1 0 0 0 1 0 0 0 0 1 0 0 0 1 0 0 0 0 0 0 0 0 1 0 0 0 1 0 0 0 0 0 1 1 0 0 0 0 0 0 0 1 1 placenta 3 middle ear bones synapsid opening long arms 3-toed foot hole in hip socket palatal opening amniote egg limbs jaws Lamprey (outgroup) therapsid (extinct) Tyrannosaurus rex (extinct) trout frog turtle sparrow kangaroo mouse antorbitol opening In this exercise you will perform a cladistic analysis of jawed vertebrates, including both living and extinct forms. Notice that this analysis includes the jawless lamprey as an outgroup. Examine the following data matrix in which a character is scored “1” if present and “0” if absent: 0 0 0 0 0 0 0 0 1 8. Create and save a file in PAST that contains the information in the table you’ve just examined. [Rename rows and columns. Note that PAST does not accept spaces, so use abbreviations, if necessary.] 9. Create a cladogram. Select all data by clicking the gray button in the upper left corner of the data matrix. Choose “Parsimony analysis” under the Cladistics menu. Use the “Exhaustive” algorithm; “Fitch” optimization; 5 reorderings; 0 Bootstrap replicates). Click the “Go” button to produce the cladogram. How many MPTs did the program find? _______________________ What is the Tree Length of this (these) MPT(s)? _______________________ How many trees were evaluated? _______________________ 10. Print a copy of the cladogram. 11. On the printed copy of the cladogram use short horizontal lines to indicate the origin of the various derived characters. Make sure to label each character. [Notice that the tree window in PAST allows you to view the distribution of characters among the taxa being investigated. You can do this by toggling up and down on the arrows next to the word “Characters” in the grey area at the right of the pop-up window.] Turn in the annotated cladogram to the instructor. Which derived character defines the clade: sparrow + turtle + T. rex? _________________________________ How does the turtle differ from the sparrow and T. rex? ____________________ _________________________________________________________________ Which derived character defines the clade: therapsid + kangaroo + mouse? _________________________________ Assignment—Part 3 (Cladistic analysis of fossil sand dollars) In this exercise you will perform cladistic analysis on ten species of fossil sand dollars using data from Mooi et al. 2000 (Journal of Paleontology, v. 74, pp. 263–281). Twentyfour characters have been selected and character-states for each character have been determined for each of the ten species. The data matrix (Table 1) is thus a 10-row × 24column array. Drawings of the ten species are given in Figure 2. 5 Table 1.—Data matrix (download file at http://faculty.cns.uni.edu/~groves/) Figure 2.—Drawings of ten sand dollar species showing variation in shape, ambulacra, and other characters. The following is a list of the twenty-four characters and their possible character-states. In each case, the “primitive” character-state is assigned the value 0 and “derived” states are assigned the value 1 or 2. 1. Test outline: 0 = circular to slightly elongate; 1 = oval 2. Test height: 0 = highly domed; 1 = flat 3. Test edge: 0 = thick and rounded; 1 = thin and sharp 4. Intestine position: 0 = not within peripheral pillars; 1 = within peripheral pillars 5. Peripheral ballast system: 0 = simple; 1 = complex 6. Ambulacral basicoronal plates: 0 = short; 1 = long 7. Interambulacral basicoronal plates: 0 = small; 1 = large 8. Interambulacral first post-basicoronal plates: 0 = short; 1 = long 9. Peristome size: 0 = small; 1 = large 10. Periproct position: 0 = not in contact with basicoronal; 1 = touching basicoronal 11. Oral interambulacral columns: 0 = discontinuous; 1 = continuous; 2 = partially continuous 12. Oral interambulacral columns: 0 = wide; 1 = narrow 13. Interambulacral columns at edge: 0 = wide; 1 = attenuated; 2 = extremely attenuated 6 14. Ambulacral indentation at edge: 0 = absent; 1 = present 15. Festooned ambulacral lunules: 0 = absent; 1 = present 16. Anal lunule: 0 = absent; 1 = short; 2 = slot-like 17. Ridge around anal lunule: 0 = absent; 1 = present 18. Ambulacral pressure drainage channels: 0 = absent; 1 = present 19. Anal lunule pressure drainage channels: 0 = absent; 1 = present 20. Food groove branching: 0 = simple; 1 = complex 21. Petaloid non-respiratory podia: 0 = present; 1 = absent 22. Number of trailing podia: 0 = many; 1 = few 23. Geniculate spine fields: 0 = absent or poorly developed; 1 = well developed 24. Locomotory spine fields: 0 = indistinct; 1 = distinct 12. Go to http://faculty.cns.uni.edu/~groves/ and save the data file titled “Sand dollars.” Open the file in PAST. Notice that species’ names have been abbreviated so that Iheringiella patagoniensis is Iher, Amplaster coloniensis is Ampc, Amplaster ellipticus is Ampe, and so on. Notice also that Proescutella is the outgroup: i.e., it does not possess any of the derived traits shared by all other species in the group. 13. Select all data in the data matrix by clicking on the gray button in the upper left corner of the matrix. Now perform cladistic analysis by choosing “Parsimony analysis” under the Cladistics menu. Use the following settings: “Branch-and-bound” algorithm; “Fitch” optimization; 5 reorderings; 0 Bootstrap replicates. Click the “Go” button to produce a cladogram. Print a copy of the cladogram to examine and turn it to the instructor. [It may be necessary to copy and paste into another application such as MS Word or MS PowerPoint.] (1) How many MPTs did the program generate? ____________ (2) What is the tree length of the MPT? ____________ 14. If everything has worked correctly, your cladogram should indicate that Monophoraster darwini and M. duboisi are more closely related to one another than either is to any other species in the group. Identify two “derived” character-states that are present in these two species but not in any other species in the group: (1) ________________________ (2) ________________________ 15. Your cladogram should indicate that the three species of Amplaster are more closely related to one another than any of the three is to any other species in the group. Identify the “derived” character-state shared only by these three species: (1) ________________________ 16. Your cladogram should indicate that Leodia sexiesperforata and Mellita quinquiesperforata are more closely related to one another than either is to any other 7 species in the group. Identify two “derived” character-states possessed by these two species but not by others: (1) ________________________ (2) ________________________ 17. According to the cladogram, Iheringiella patagoniensis is relatively distant from the branches that end in Monophoraster and Amplaster species. Examine Table 1 and notice that I. patagoniensis, Monophoraster spp. and Amplaster spp. all possess long interambulacral first post-basicoronal plates (character #8, character-state = 1). Explain how this “derived” character-state could be present in distantly related species: 8