Survival outcomes of myeloid malignancies with borderline
advertisement

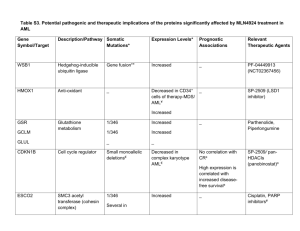
Letter to the Editor Prognosis in patients with MDS or AML and bone marrow blasts between 10% and 30% is not associated with blast counts but depends on cytogenetic and molecular genetic parameters - Online Supplement - ADDITIONAL REFERENCES RELATED TO METHODS Cytomorphology 1. Löffler H, Raststetter J, Haferlach T. Atlas of Clinical Hematology. 6th edition. Springer, Berlin, 2004. 2. Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the FrenchAmerican-British Cooperative Group. Ann Intern Med 1985; 103: 620-5. 3. Swerdlow S, Campo E, Lee Harris N, Jaffe E, Pileri S, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th edition. IARC press, Lyon, 2008. Chromosome banding analysis 4. Schoch C, Schnittger S, Bursch S, Gerstner D, Hochhaus A, Berger U, et al. Comparison of chromosome banding analysis, interphase- and hypermetaphase-FISH, qualitative and quantitative PCR for diagnosis and for follow-up in chronic myeloid leukemia: a study on 350 cases. Leukemia 2002; 16: 53-9. 1 Molecular mutation analysis 5. Schnittger S, Kern W, Tschulik C, Weiss T, Dicker F, Falini B, et al. Minimal residual disease levels assessed by NPM1 mutation specific RQ-PCR provide important prognostic information in AML. Blood 2009; 114: 2220-31. 6. Schnittger S, Schoch C, Dugas M, Kern W, Staib P, Wuchter C, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood 2002; 100: 59-66. 7. Weisser M, Kern W, Schoch C, Hiddemann W, Haferlach T, Schnittger S. Risk assessment by monitoring expression levels of partial tandem duplications in the MLL gene in acute myeloid leukemia during therapy. Haematologica 2005; 90: 881-9. 8. Bacher U, Haferlach C, Kern W, Haferlach T, Schnittger S. Prognostic relevance of FLT3-TKD mutations in AML: the combination matters--an analysis of 3082 patients. Blood 2008; 111: 2527-37. 9. Bacher U, Haferlach T, Schoch C, Kern W, Schnittger S. Implications of NRAS mutations in AML: a study of 2502 patients. Blood 2006; 107: 3847-53. 10. Snaddon J, Smith ML, Neat M, Cambal-Parrales M, Dixon-McIver A, Arch R, et al. Mutations of CEBPA in acute myeloid leukemia FAB types M1 and M2. Genes Chromosomes Cancer 2003; 37: 72-8. 11. Schnittger S, Dicker F, Kern W, Wendland N, Sundermann J, Alpermann T, et al. RUNX1 mutations are frequent in de novo AML with non complex karyotype and confer an unfavourable prognosis. Blood 2010; epub ahead of print 9 Dec 2010; doi: 10.1182/blood2009-11-255976. 12. Schnittger S, Haferlach C, Ulke M, Alpermann T, Kern W, Haferlach T. IDH1 mutations are detected in 6.6% of 1414 AML patients and are associated with intermediate risk karyotype and unfavorable prognosis in adults younger than 60 years and unmutated NPM1 status. Blood 2010; 116: 5486-96. 2 Immunophenotyping with multiparameter flow cytometry (MFC) 13. Kern W, Voskova D, Schoch C, Hiddemann W, Schnittger S, Haferlach T. Determination of relapse risk based on assessment of minimal residual disease during complete remission by multiparameter flow cytometry in unselected patients with acute myeloid leukemia. Blood 2004; 104: 3078-85. 14. Kern W, Haferlach C, Schnittger S, Haferlach T. Clinical utility of multiparameter flow cytometry in the diagnosis of 1013 patients with suspected myelodysplastic syndrome: correlation to cytomorphology, cytogenetics, and clinical data. Cancer 2010; 116: 4549-63. Microarray gene expression profiling 15. Kohlmann A, Kipps TJ, Rassenti LZ, Downing JR, Shurtleff SA, Mills KI, et al. An international standardization programme towards the application of gene expression profiling in routine leukaemia diagnostics: the Microarray Innovations in LEukemia study prephase. Br J Haematol 2008; 142: 802-7. 16. Kohlmann A, Haschke-Becher E, Wimmer B, Huber-Wechselberger A, Meyer-Monard S, Huxol H, et al. Intraplatform reproducibility and technical precision of gene expression profiling in 4 laboratories investigating 160 leukemia samples: the DACH study. Clin Chem 2008; 54: 1705-15. 17. Miesner M, Haferlach C, Bacher U, Weiss T, Macijewski K, Kohlmann A, Klein HU, Dugas M, Kern W, Schnittger S, Haferlach T. Multilineage dysplasia (MLD) in acute myeloid leukemia (AML) correlates with MDS-related cytogenetic abnormalities and a prior history of MDS or MDS/MPN but has no independent prognostic relevance: a comparison of 408 cases classified as "AML not otherwise specified" (AML-NOS) or "AML with myelodysplasia-related changes" (AML-MRC). Blood 2010; 116: 2742-51. 3 SUPPLEMENTARY TABLES Table S1: Overall survival (OS) according to cytomorphology, cytogenetics, and molecular genetics. Parameter Number of patients Median OS (months) 10%-19% BM blasts (= MDS cohort) 20%-30% (= AML cohort) 96 180 34.4 43.7 Cytogenetic risk group (MRC criteria) intermediate karyotype p Separation in two blast categories n.s. 213 43.7 unfavorable karyotype Molecular subgroups 63 13.9 NPM1 mutated NPM1 wild-type 50 226 n.r. 27.3 0.005 FLT3-ITD positive FLT3-ITD negative 12 264 34.9 14.7 n.s. NPM1-mutated/FLT3-ITD-negative all other NPM1/FLT3-ITD compositions 41 235 n.r. 27.3 0.002 MLL-PTD positive MLL-PTD negative 18 258 34.9 n.r. n.s. RUNX1-mutated RUNX1 wild-type 34 82 14.1 n.r. n.s. 4 <0.001 Table S2: Uni- and multivariate analyses for definition of prognostically relevant parameters with regard to overall survival (OS). *Cytogenetic risk stratification followed revised MRC criteria. **NPM1mut/FLT3-ITD-negative status was associated with a better prognosis when compared to all other molecular subgroups. Due to overlapping features, only NPM1mut/FLT3-ITD-negative status, but not the NPM1mut status considered as single parameter, was included in the multivariate analysis. Parameter Univariate (p) Multivariate (p) age WBC count <0.001 n.s. 0.048 - platelets hemoglobin 0.018 n.s. 0.033 - 10%-19% vs 20%-30% BM blasts cytogenetic risk group* n.s. <0.001 0.010 FLT3-ITD mutation status alone NPM1mut status alone n.s. 0.008 - NPM1mut/FLT3ITD-negative status vs all others** 0.005 n.s. MLL-PTDmut status alone RUNX1mut status alone n.s. n.s. - CD34+ cells (multiparameter flow cytometry) n.s. - 5 Legend to Supplemental Table S3A-C Table S3: Results of gene expression analysis from 26 patients of the cohort. A: comparing the MDS (n=11) versus AML (n=15) cases; B: comparing cases with intermediate (n=16) and with unfavorable karyotypes (n=10); C: comparing cases with normal (n=9) and abnormal (n=17) karyotypes. 6 SUPPLEMENTARY FIGURES Supplemental Figure S1: Frequency of cytogenetic and molecular alterations in MDS and AML cohorts. The y-axis shows the percentages of cases carrying the respective genetic alteration. The grey columns visualize the MDS cases, the red columns the AML patients, respectively (unfav.: unfavorable; KTs: karyotypes; complex: complex aberrant karyotypes). Supplemental Figure S2: Hierarchical clustering analysis of global gene expression profiles. For supervised statistical analyses of the gene expression data, the samples were grouped accordingly and for each respective category differentially expressed genes were calculated by means of t-tests. To assess significance, false discovery rates (FDR) were calculated. Results of gene expression profiling are shown in a total of 26 cases (15 AML, 11 MDS) by hierarchical clustering using Euclidean distance and complete linkage parameters. The 26 cases are shown in columns; the rows depict the gene expression intensity for the genes from the top differentially expressed genes. Each heatmap depicts the gene expression signature for the top-500 differentially detected probe sets. A: Results of gene expression analysis comparing cases with 10%-19% (group 0) and cases with 20%-30% of BM blasts (group 1). B: Gene expression profiles of cases with intermediate (group 1) versus unfavorable karyotypes (group 2). C: Gene expression profiles of cases with normal (group 0) versus abnormal karyotypes (group 1). Detailed information on the differentially expressed probe sets and their functional annotation is available online. Note: the raw p-value did not remain significant after strict adjustment for multiple testing. 7