Supplementary Methods - Word file (40 KB )
advertisement

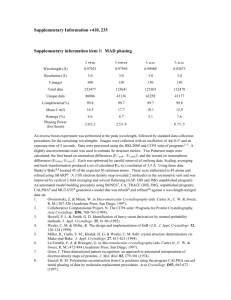
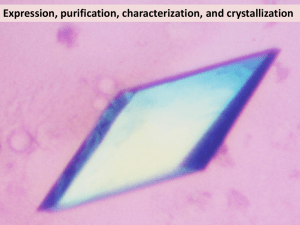
Supplementary information Methods Over-expression and purification of NhaA Cells of the E. coli strain RK201 transformed with the plasmids pAXH encoding His-tagged NhaA2 and pIQ (a pACYC184 derivative) encoding LacIQ were used for over-expression of NhaA. The gene nhaA was deleted from RK20, a property that was found essential for the purification of NhaA. Protein was produced3 in RK20/pAXH/pIQ cells after induction (at A600 = 0.6) with 0.5 mM isopropyl-β-D-thiogalactopyranoside for 2 h at 37 ºC. Cells were harvested and high pressure membranes3 were prepared and frozen (10 mg membrane protein/ml) in liquid nitrogen and stored at –80 ºC. Protein was extracted from the membranes at 4 ºC for 1 h using a solution containing 1 % ndodecyl-β-D-maltopyranoside, 100 mM MOPS/KOH (pH 7), 28% glycerol, 3 mM Tris/Cl, 83 mM sucrose and 47 mM choline chloride. The protein was bound to a Ni2+NTA affinity column and purified as described3. On the column, the detergent was exchanged to 0.03 % n-dodecyl-α-D-maltopyranoside in 20 mM Bis-Tris (pH 6.5). Elution was conducted at acidic pH (25 mM Na+/citrate, 5 mM MgCl2, 100 mM KCl, pH 4). The protein was concentrated (Centriprep YM-50) to 12 mg/ml and frozen in liquid nitrogen in the above solution supplemented with 20 % sucrose and stored at –80 ºC. For protein labelled with seleno-L-methionine, E. coli B834DE39 (Novagen, WI, USA) transformed with pAXH and pIQ were grown in minimal medium (L-methionine 50 µg/ml replaced by seleno-L-methionine) and the protein was processed as above. Crystallisation Crystals were grown by hanging-drop vapour diffusion at 6 ºC by mixing equal volumes of protein (4-6 mg/ml) and reservoir solutions. NhaA crystals were grown over 1 a reservoir solution of 28-34% polyethylene glycol 400, 200-450 mM MgCl2, 100 mM KCl, 25 mM Na+-citrate (pH 4), 1% n-octyl-β-D-glucopyranoside and 0.5 % ethanol. Crystals appeared in 4 days and reached a maximal size of 0.2 x 0.3 x 0.3 mm in 15 days. The crystals were gently removed from the drop and flash frozen with nitrogen (100 K). Data collection and structure determination Diffraction data were collected at the ESRF beam lines ID29 (data set SeMet SAD) and ID23 (data set Native 1) and at the SLS beam line PX06 (data set Native 2) with charge-coupled device detectors and cryo-cooling (100 K). Data were processed and scaled using the programs HKL4, XDS5 and the CCP4 program suites6. The crystals belong to an orthorombic space group P212121. Two molecules NhaA are present in the asymmetric unit. The structure was solved by SAD technique using 20 selenium atoms. Initial positions were found using Solve7 and were refined with SHARP8. Phases were calculated to 4.3 Å (17 - 4.3 Å) using SHARP with a figure of merit of 0.42 for acentric reflections and 0.13 for centric reflections, a phasing power (anomalous) of 2.4 and Rcullis (anomalous) of 0.52 for acentric reflections. The SAD phases were applied to the native data set (native1) and gradually extended to 3.8 Å using the program DM 9 with solvent flattening, histogram matching and averaging options. The correlation between the electron density areas related by non-crystallographic symmetry was 78.8 % after density modification. An initial model was built using the program O10 and refinement was carried out using CNS11 with tight two-fold NCS restraints. For final refinement of the model the second native data set at 3.45 Å was used. 2 Fig. 1 and 3b were prepared using the program O, Fig. 2, 4, S1 and S2 were made with MOLSCRIPT12 and rendered with RASTER3D13. Fig. 3a was prepared using Pymol14 and Fig. 3b-d with the program GRASP15 References Supplementary Information 1. Padan, E., Maisler, N., Taglicht, D., Karpel, R. & Schuldiner, S. Deletion of ant in Escherichia coli reveals its function in adaptation to high salinity and an alternative Na+/H+ antiporter system(s). J. Biol. Chem. 264, 20297-20302 (1989). 2. Olami, Y., Rimon, A., Gerchman, Y., Rothman, A. & Padan, E. Histidine 225, a residue of the NhaA-Na+/H++ antiporter of Escherichia coli is exposed and faces the cell exterior. J. Biol. Chem. 272, 1761-1768 (1997). 3. Venturi, M. & Padan, E. Purification of NhaA Na+/H+ antiporter of Escherichia coli for 3D or 2D crystallization. in A Practical Guide to Membrane Protein Purification (eds. Hunte, C., Von Jagow, G. & Schagger, H.) 179-190 (Academic Press, Amsterdam, 2002). 4. Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collection in oscillation mode. Methods Enzymol. 276, 307-326 (1997). 5. Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795800 (1993). 6. Collaborative Computer Project No. 4. The CCP4 Suit: Programs for Protein Crystallography. Acta Cryst. D50, 760-763 (1994). 7. Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D55, 849-861 (1999). 8. de la Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol 276, 472-494 (1997). 9. Cowtan, K. D. Phase combination and cross validation in iterated densitymodification calculations. Acta Crystallogr. D52, 43-48 (1996). 10. Jones, T. A. & Kjeldgaad, M. Electron density map interpretation. Methods Enzymol. 277, 173-208 (1997). 11. Brunger, A. T. et al. Crystallography & NMR System. Acta Crystallogr. D54, 905-921 (1998). 12. Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946-950 (1991). 13. Merritt, E. A. & Murphy, M. E. P. Raster3D 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D50, 869-873 (1994). 14. http://pymol.sourceforge.net/. 15. Honig, B. & Nicholls, A. Classical electrostatics in biology and chemistry. Science 268, 1144-1149 (1995). 16. http://www.ebi.ac.uk/dali/ 3