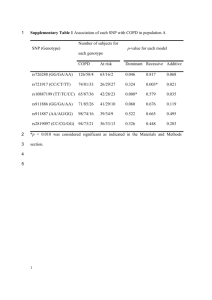
Supplementary Figure 1: Plot of estimated relative risks (RRs) from
advertisement
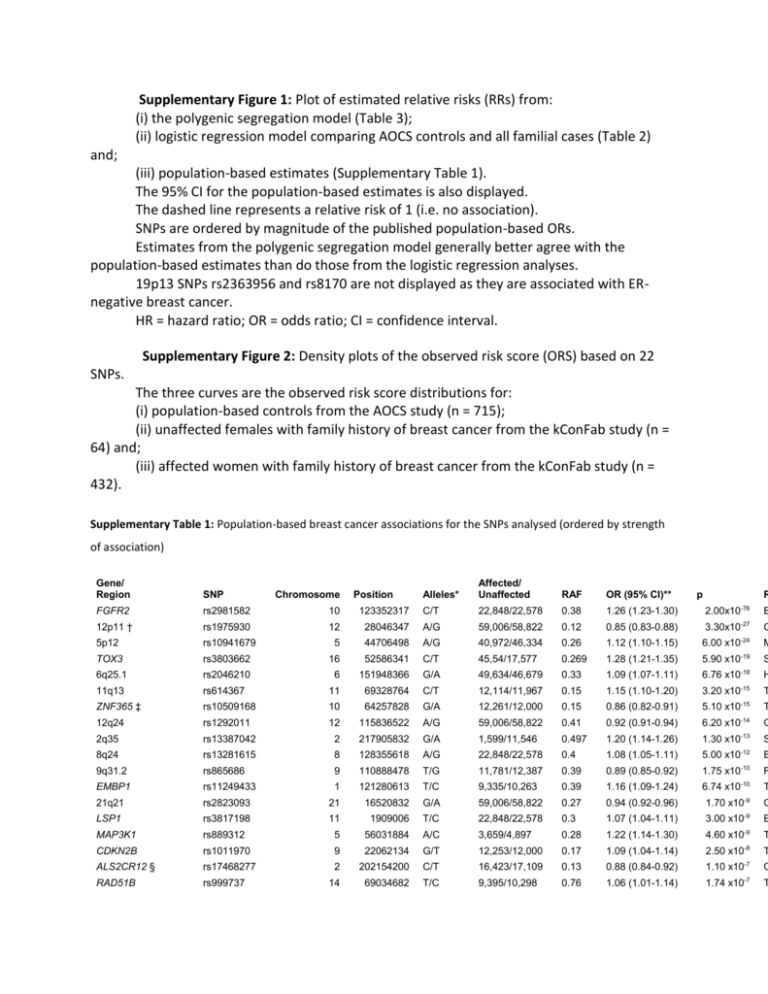
Supplementary Figure 1: Plot of estimated relative risks (RRs) from: (i) the polygenic segregation model (Table 3); (ii) logistic regression model comparing AOCS controls and all familial cases (Table 2) and; (iii) population-based estimates (Supplementary Table 1). The 95% CI for the population-based estimates is also displayed. The dashed line represents a relative risk of 1 (i.e. no association). SNPs are ordered by magnitude of the published population-based ORs. Estimates from the polygenic segregation model generally better agree with the population-based estimates than do those from the logistic regression analyses. 19p13 SNPs rs2363956 and rs8170 are not displayed as they are associated with ERnegative breast cancer. HR = hazard ratio; OR = odds ratio; CI = confidence interval. Supplementary Figure 2: Density plots of the observed risk score (ORS) based on 22 SNPs. The three curves are the observed risk score distributions for: (i) population-based controls from the AOCS study (n = 715); (ii) unaffected females with family history of breast cancer from the kConFab study (n = 64) and; (iii) affected women with family history of breast cancer from the kConFab study (n = 432). Supplementary Table 1: Population-based breast cancer associations for the SNPs analysed (ordered by strength of association) Gene/ Region SNP Alleles* Affected/ Unaffected FGFR2 rs2981582 10 RAF OR (95% CI)** 123352317 C/T 22,848/22,578 0.38 1.26 (1.23-1.30) 2.00x10-76 12p11 † rs1975930 E 12 28046347 A/G 59,006/58,822 0.12 0.85 (0.83-0.88) 3.30x10-27 5p12 rs10941679 G 5 44706498 A/G 40,972/46,334 0.26 1.12 (1.10-1.15) 6.00 x10-24 M -19 S Chromosome Position p R TOX3 rs3803662 16 52586341 C/T 45,54/17,577 0.269 1.28 (1.21-1.35) 5.90 x10 6q25.1 rs2046210 6 151948366 G/A 49,634/46,679 0.33 1.09 (1.07-1.11) 6.76 x10-18 H 11q13 rs614367 11 69328764 C/T 12,114/11,967 0.15 1.15 (1.10-1.20) 3.20 x10-15 T ZNF365 ‡ rs10509168 10 64257828 G/A 12,261/12,000 0.15 0.86 (0.82-0.91) 5.10 x10-15 T 12q24 rs1292011 12 115836522 A/G 59,006/58,822 0.41 0.92 (0.91-0.94) 6.20 x10-14 G 2q35 rs13387042 2 217905832 G/A 1,599/11,546 0.497 1.20 (1.14-1.26) 1.30 x10-13 S -12 E 8q24 rs13281615 8 128355618 A/G 22,848/22,578 0.4 1.08 (1.05-1.11) 5.00 x10 9q31.2 rs865686 9 110888478 T/G 11,781/12,387 0.39 0.89 (0.85-0.92) 1.75 x10-10 F EMBP1 rs11249433 1 121280613 T/C 9,335/10,263 0.39 1.16 (1.09-1.24) 6.74 x10-10 T 21q21 rs2823093 21 16520832 G/A 59,006/58,822 0.27 0.94 (0.92-0.96) 1.70 x10-9 G LSP1 rs3817198 11 1909006 T/C 22,848/22,578 0.3 1.07 (1.04-1.11) 3.00 x10-9 E -9 T MAP3K1 rs889312 5 56031884 A/C 3,659/4,897 0.28 1.22 (1.14-1.30) 4.60 x10 CDKN2B rs1011970 9 22062134 G/T 12,253/12,000 0.17 1.09 (1.04-1.14) 2.50 x10-8 T ALS2CR12 § rs17468277 2 202154200 C/T 16,423/17,109 0.13 0.88 (0.84-0.92) 1.10 x10-7 C RAD51B rs999737 14 69034682 T/C 9,395/10,298 0.76 1.06 (1.01-1.14) 1.74 x10-7 T ANKRD16/FBXO18 rs2380205 10 5886734 C/T 12,235/11,961 0.43 0.94 (0.91-0.98) 4.60 x10-7 T SLC4A7 rs4973768 3 27416013 C/T 3,659/4,897 0.47 1.16 (1.10-1.24) 5.80 x10-7 T STXBP4 rs6504950 17 53056471 G/A 30,470/33,302 0.27 0.95 (0.92-0.97) 0.0001 A 19p13 ¶ rs2363956 19 17394124 A/C 833/6,603 0.507 0.83 (0.75-0.92) 0.0003 A 19p13 ¶ rs8170 19 17389704 G/A 819/6,516 0.189 1.21 (1.07-1.37) 0.0029 A C6orf97 rs12662670 6 151918856 T/G 7,899/11,234 0.071 1.12 (1.03-1.21) 0.0064 S RAF = risk allele frequency; OR = odds ratio; CI = confidence interval * Non-risk/risk allele ** All ORs are per-allele unless otherwise stated † Proxy SNP rs10771399 (r2 = 0.95) was reported; ‡ Proxy SNP ZNF365 rs10995190 was reported; § rs17468277 is a proxy for SNP for SNP CASP8 D302H rs1045485 (r2 = 1.00); ¶ ER-negative breast cancer association reported Supplementary Table 2: Meta-analysis pooling parent-of-origin associations for SNP rs3817198 in the LSP1 gene Maternal effect Paternal effect Study Kong et al kConFab/AOCS Meta-analysis Unaffected/ affected 34,909/1,803 3,588/1,463 38,497/3,266 β -0.0943 -0.0580 -0.0766 σ 0.0582 0.0595 0.0416 Weight (%) 51.09 48.91 RR (95% CI) 0.91 (0.81-1.02) 0.94 (0.84-1.06) 0.93 (0.85-1.01) p 0.11 0.33 0.07 Kong et al kConFab/AOCS Meta-analysis 34,909/1,803 3,588/1,463 38,497/3,266 0.1570 0.1119 0.1384 0.0538 0.0641 0.0412 58.72 41.28 1.17 (1.05-1.30) 1.12 (0.99-1.27) 1.15 (1.06-1.24) 0.0038 0.0810* 0.0008 RR = relative risk; CI = confidence interval; β = estimated log-RR; σ = estimated standard error for the log-RR RR is an odds ratio (OR) in the case of Kong et al and is a hazard ratio (HR) for the kConFab/AOCS analysis Weights were calculated by inverse variance weighting The reported effect allele for SNP rs3817198 was the C allele (other allele = T). * p-value when testing HR > 1 was 0.040