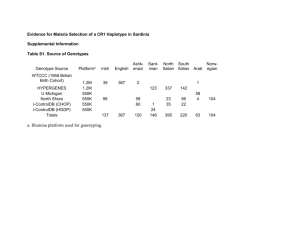
Supplement 1
advertisement
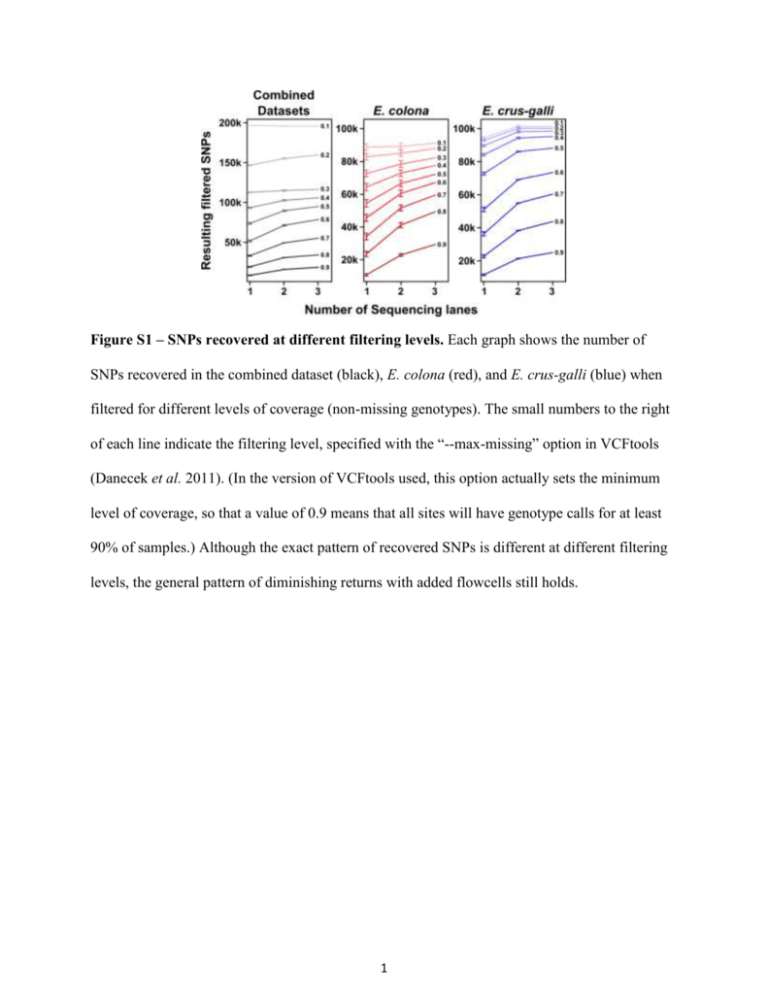
Figure S1 – SNPs recovered at different filtering levels. Each graph shows the number of SNPs recovered in the combined dataset (black), E. colona (red), and E. crus-galli (blue) when filtered for different levels of coverage (non-missing genotypes). The small numbers to the right of each line indicate the filtering level, specified with the “--max-missing” option in VCFtools (Danecek et al. 2011). (In the version of VCFtools used, this option actually sets the minimum level of coverage, so that a value of 0.9 means that all sites will have genotype calls for at least 90% of samples.) Although the exact pattern of recovered SNPs is different at different filtering levels, the general pattern of diminishing returns with added flowcells still holds. 1 Figure S2 – Allele depth ratios. Allele sequencing depth was tabulated across all sites with at least 6 reads contributing to the genotype. The graph shows a histogram of the proportion of total allele depth belonging to the minor allele; “minor allele” in this case refers to which allele was less common in that specific sample, not across the population as a whole. Since both species of barnyard millet are hexaploid (Prasada Rao et al. 1993, Upadhyaya et al. 2008), in theory the depths should cluster around 0.16, 0.33, and 0.5 (corresponding to 1, 2, or 3 out of 6 copies). Although there are spikes around 0.33 and 0.5, there is still a large amount of spread across the rest of the range. Trying to assign copy numbers based off these data would be risky at best, which is why in our analyses we instead use the discriminating SNP dataset. 2 Figure S3 – Population structure and phylogeny in the complete SNP dataset. The same population structure and phylogenetic analyses that were used to generate Fig. 3 (main text) were also applied to the entire, non-discriminating SNP dataset. Although the results for the combined set and for E. crus-galli are qualitatively similar to those with the discriminating SNP set, the presence of homeologous SNPs makes it difficult to identify any meaningful structure within E. colona. 3 4 Figure S4 – Population structure at different clustering levels for the discriminating SNP set. Population assignments for each individual are shown for the entire collection (left) and for the two species separately (middle and right). The number of clusters (K) was varied from 2 to 10, and the one chosen for display in the main text is marked by a black box. 5 Figure S5 – Phylogenetic webs for each dataset. The full phylogenetic webs from SplitsTree4 are shown for each dataset, with labels colored according to Fig. 3 (main text). 6 Figure S6 – Species and country assignations for the entire collection. The phylogenetic webs for the entire collection are shown. Accession names are colored according to the species name (a) or country of origin (b) listed in the accessions' passport data. Mismatches for the species names may be due to misclassification or to simple labeling errors at some point during sample preparation (seed storage, grow-out, DNA prep, etc.). 7 Figure S7 – Race and cluster assignments within each species. The phylogenetic webs for each species are shown, with labels indicating the assigned race (a) or morphological cluster (b) within each species. (a) Of the races, only E. crus-galli race Crus-galli appears to consistently cluster by phylogeny. The racial assignments of all other groups appear to be independent of phylogenetics. (b) The morphological clusters used to create the core collection (Upadhyaya et al. 2014) also do not generally match up with phylogeny. 8