Zoo/Bot 3333 Genetics Quiz 4 11/18/12 For answers to the quiz
advertisement

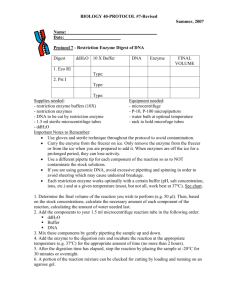
Zoo/Bot 3333 Genetics Quiz 4 11/18/12 For answers to the quiz, click here Questions 1-3 pertain to the following. The size of the E. coli genome is about 3.2 x 106 bp. 1. Approximately how long is the bacterial chromosome? a) 0.5 μm; b) 3 μm; c) 200 μm; d) 800 μm; e) 1,100 μm. 2. If the E. coli chromosome completes one round of replication in 60 minutes, about how fast is the DNA unwinding at a replication fork, in terms of revolutions per minute? (hint: review how E. coli replicates!) a) 62 RPM; b) 200 RPM; c) 2,700 RPM; d) 53,000 RPM; e) 2 million RPM. 3. The BamHI restriction endonuclease cuts at the following sequence: 5’-GGATC C-3’ 3’-C CTAGG-5’ Assuming that the genome contains equal proportions of all four nucleotides, and their distribution is random, approximately how many fragments would be produced on BamHI digestion of the E. coli genome? a) 28; b) 780; c) 5.3 x 104; d) 7.3 x 105; e) 1.9 x 107. 4. The restriction enzyme, AvaI has the six base recognition sequence CPyCGPuG, where Py stands for a pyrimidine, and Pu stands for a purine). On average, this enzyme might be expected to cut a random DNA sample every ____________bp. a) 64; b) 256; c) 1024; d) 1296; e) 4096. 5. The recognition sites for the restriction enzymes BamHI, XbaI and BglII are, respectively: BamHI: 5’-GGATC C-3’ 3’-C CTAGG-5’ XbaI: 5’-TCTAG A-3’ 3’-A GATCT-5’ BglII: 5’-AGATC T-3’ 3’-T CTAGA-5’ where the arrows represent the cut locations for each strand of the palindromic site. Which enzymes leave compatible ends that will facilitate ligation? a) All of these enzymes leave ends that are compatible with ends generated by the others; b) None of the enzymes produce compatible ends; c) Only BamHI and BglII fragments are compatible; d) Only BamHI and XbaI fragments are compatible; e) only BglII and XbaI fragments are compatible. Questions 6-8 pertain to the following. A mouse null mutant for a particular enzyme has been identified by the fact that heterozygotes for the mutation produce ½ the amount of enzyme as normal mice. By comparing mutant to wild type proteins on electrophoretic gels, the enzyme protein is identified, eventually leading to the biochemical isolation of the enzyme protein. A small portion of the sequence of the purified enzyme protein is determined to be: Met Gly Ser Ala Gln Tyr Met Phe Asp Glu Trp Ser Met Ile Leu This information is used to synthesize 21 base ‘degenerate’ oligonucleotides that will be used to screen a cDNA library by nucleic acid hybridization for the cDNA encoding this particular enzyme. 6. What region of the amino acid sequence above should be used to manufacture an appropriate ‘degenerate’ probe? a) Met-Gly-Ser-Ala-Gln-Tyr-Met; b) Ser-Ala-Gln-Tyr-Met-Phe-Asp; c) Gln-Tyr-Met-Phe-Asp-Glu-Trp; d) Asp-Glu-Trp-Ser-Met-Ile-Leu; e) any of the above would be equally effective. 7. How many different oligonucleotides would be present in the above degenerate probe? a) 1; b) 7; c) 12; d) 32; e) none of the above. 8. True or false. A single stranded degenerate probe encoding the protein sequence indicated above could be also be used for a Northern blot analysis. Questions 9-10 pertain to the above: A large family is segregating an autosomal dominant disease of late onset (approximately 40 years of age). A genomic DNA sample from each family member is digested with the restriction enzyme BamHI run on an electrophoretic gel, and subjected to Southern blot analysis. The probe used in this instance hybridizes to a DNA fragment linked to the disease gene, which shows polymorphism for this restriction enzyme. The autoradiogram of this blot is shown above, aligned with the family pedigree. 9. In the above example, which restriction fragments are on the same chromosome homolog as the disease gene allele in individual I-1? a) the 2 kb fragment; b) the 8 kb fragment; c) both the 2 and 8 kb fragments; d) the 10 kb fragment; e) all three of the above fragments. 10. Which of the following most accurately represents the map distance between the disease gene and the marker (probe DNA) locus based on this pedigree? a) 3.6 map units; b) 7.1 map units; c) 10.7 map units; d) 14.3 map units; e) 21.4 map units.