Name LAB REPORT 2: Enzyme Interaction With DNA 40 points 1 (5
advertisement

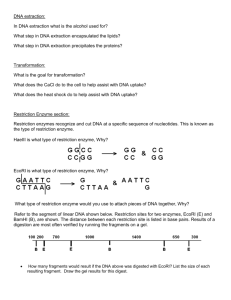
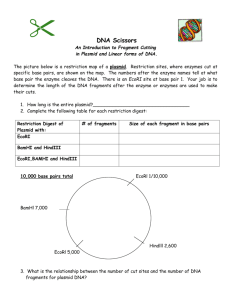
Name LAB REPORT 2: Enzyme Interaction With DNA 40 points 1 (5) What is the difference between an endonuclease and an exonuclease? What effect would you expect each to have on cut and uncut pRHR53? Why? 2 (5) What is the difference between a site-specific endonuclease and a general endonuclease? How would you expect the lanes on a gel to differ if you cut the same DNA with each type of enzyme? Why? 3 (5) What is the concentration of pRHR53 (consult the syllabus or GE web page)? How many g of DNA did you use for each reaction? 4 (10) Write the Methods and Materials for Experiment 5. 5 (10) Attach a copy of the gel for Experiment 5. 6 (30) Analyze the results of your gel by completing the table. For each enzyme use a (+) or (--) to indicate whether it acts on each substrate. In the last column, briefly describe the appearance of the lane and explain the result. Enzyme Circular Linear Circular SS Linear SS Appearance of Lane & Explanation No Enzyme HindIII DNase I exonuclease T5 exonuclease Mung Bean nuclease 7 (5) Complete the table using information in Appendix 10 or the New England Biolabs catalog. Enzyme Conc (U/ml) # Units per rxn Unit Definition BglII HindIII DNase I exonuclease T5 exonuclease Mung Bean nuclease* *stock was diluted 1/10 8 (10) Write the Methods and Materials for Experiment 6. 9 (10) Attach a copy of the gel for Experiment 6 10 (30) Analyze the results of your gel by completing the table. For each substrate check the appropriate box to indicate whether or not it was cut by each REase. Substrate REase none DNA treated with M.EcoRI R.EcoRI R.BamHI none DNA treated with M.BamHI R.EcoRI M.BamHI Cut Uncut Explanation 11 (20) Complete the table using information in Appendix 10 or the New England Biolabs catalog. Indicate where the cuts are made and use an asterisk to indicate the base that is modified by the MTase. Enzyme Recognition Sequence Conc (U/ml) Unit Definition M.EcoRI R.EcoRI M.BamHI R.BamHI 12 (5) What is the concentration of DNA (consult Appendix 10)? How many g of DNA did you use in each reaction? 13 (5) Draw a diagram of S-adenosyl methionine. Grade = Total Points x 40 points 150