Supplemental Material
advertisement

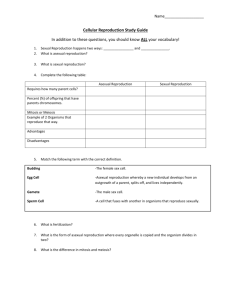
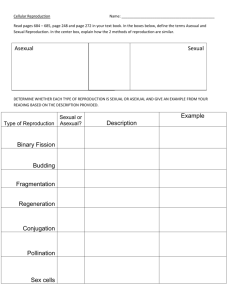
Table 1: Species generation times used in this study Generation Reference Comments time used Anopheles gambiae (aga) 10 days 1 Ranges from 5-14 days Egg to emergence of Drones is 24 days Apis mellifera (ame) 40 days 2 and it reaches sexual maturity 12-14 days after emergence Arabidopsis thaliana (ath) 45 days 3 Gestation period of 9 months + 1 year to Bos taurus (bta) 2 years 4 sexual maturity Caenorhabditis elegans (cel) 3 days 3 Gestation period of 2 months + 6-12 Canis familiaris (cfa) 11 months 4 months to sexual maturity Danio rerio (dre) 3 months 3 Asexual reproduction is 4 hrs. Sexual reproduction (spore-spore) - 3 days. However asexual reproduction is Dictyostelium discoideum (ddi) 8 hrs 3,5 common and sexual reproduction occurs only upon starvation (See ref 5). Four hours was chosen arbitrarily by giving more weight to asexual reproduction. Drosophila melanogaster (dme) 12 days 3 Entamoeba histolytica (ehi) 10 hrs 6 Gallus gallus (gga) 5 months 3 Homo sapiens (hsa) 20 years 7 Gestation period of 21 days + 5-7 weeks Mus musculus (mmu) 65 days 4 to sexual maturity Crop duration for rice varieties ranges Oryza sativa (osa) 4.5 months 8 from 3 - 9 months Saccharomyces cerevisiae (sce) 2.5 hrs 9 Strongylocentrotus purpuratus(spu) 1 year 3 Asexual reproduction is the predominant mode of reproduction and takes about 1.5 hrs. Sexual reproduction from zygote to zygote Tetrahymena thermophila (tth) 3 hrs 3, 10 ranges from 1day to14 days and this occurs once in ~ 100 generations See ref 10. Three hours was chosen arbitrarily by giving more weight to asexual reproduction. Tribolium castaneum (tca) 70 days 11 Ranges from 7-12 weeks Trypanosoma cruzi (tcr) 1 day 12 Xenopus tropicalis (xtr) 4 months 13 Species (abbreviation) Figure 1. Neighbor-joining tree was constructed using 32 orthologous protein coding genes common to all 20 species. The orthology was determined by reciprocal BLAST searches of the proteomes. The tree was constructed using Poisson corrected distance. The topology and branch lengths were similar when different gamma parameters were used. This tree topology and branch lengths were used for independent contrast analysis using the CONTRAST package of PHYLIP (14). Figure 2. This tree is based on previously published phylogenies of mammals (15), insects (16), coelamates (17) and basal eukaryotes (18). The relationships of the other clades are well known (see, Tree of life web project, http://www.tolweb.org). The polytomies are due to the uncertain relationship among basal eukaryotes (18). The independent contrast analysis was conducted using the software CAIC (19). This program assumes that the ages of taxa are proportional to the number of species they contain and uses an algorithm from Grafen (20) to obtain the relative length of each branch. Table 2. Results of independent Contrast Analysis Contrast 1 Contrast 2 No. of contrasts Slope Pearson r P - value PHYLIP Gen. time a ENC' ENC' Gen. time 19 19 -0.15 -3.39 -0.72 -0.72 0.0003 0.0003 CAIC Gen. time ENC' ENC' Gen. time 15 15 -0.2 -2.97 -0.78 -0.78 0.0004 0.0004 ENC' Std.Devb 15 -0.28 0.10 Gen. time Std.Dev 15 1.2 0.11 c Gen. time Std.Dev 15 1.4 0.23 a – Log transformed values of generation time and ENC' estimates. b – Square root of the sum of the branch lengths in a given node. c – After excluding two outliers. 0.71 0.69 0.39 References 1. Center for disease control and prevention, USA Link: http://www.cdc.gov/malaria/biology/mosquito/ 2. British bee keepers’ Association, UK Link: http://www.bbka.org.uk/articles/life_cycle_apis_mellifera.php) 3. National Institute of Health, USA Links: Arabidopsis - http://www.nih.gov/science/models/arabidopsis/index.html Worm, fruit fly and Zebra fish - http://www.nih.gov/science/models/nmm/appc1.html Dictyostellium and Tetrahymena - http://www.nih.gov/science/models/nmm/appc2.html Chicken - http://www.nih.gov/science/models/nmm/appc3.html Sea urchin - http://www.nih.gov/science/models/nmm/appc4.html 4. Animal Diversity web of University of Michigan, USA Link: http://animaldiversity.ummz.umich.edu/ 5. DictyBase, Northwestern University, USA Link: http://dictybase.org/tutorial/about_dictyostelium.htm 6. VOIGT, H., J. C. OLIVO, P. SANSONETTI and N. GUILLEN, 1999. Myosin IB from Entamoeba histolytica is involved in phagocytosis of human erythrocytes. Journal of Cell Science 112: 1191-1201. 7. NACHMAN, M. W., and S. L. CROWELL, 2000. Estimate of the mutation rate per nucleotide in humans. Genetics 156: 297-304. 8. International Rice Research Institute, Philippines Link: http://www.irri.org/ 9. EAGLESTONE, S. S., L. W. RUDDOCK, B. S. COX and M. F. TUITE, 2000. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI+] of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America 97: 240-244. 10. Tetrahymena genome database, Stanford University, USA Link: http://www.ciliate.org/genetics.shtml#Tetrahymena 11. Ohio State University, USA Link: http://ohioline.osu.edu/hyg-fact/2000/2087.html 12. VILLARREAL, D., P. NIRDE, M. HIDE, C. BARNABE and M. TIBAYRENC, 2005. Differential gene expression in benznidazole-resistant Trypanosoma cruzi parasites. Antimicrobial Agents and Chemotherapy 49: 2701-2709. 13. Sanger Institute, UK Link: http://www.sanger.ac.uk/Projects/X_tropicalis/ 14. FELSENSTEIN, J., 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington, Seattle. 15. MURPHY, W. J., E. EIZIRIK, S. J. O'BRIEN, O. MADSEN, M. SCALLY et al., 2001. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294: 2348-2351. 16. SAVARD, J., D. TAUTZ, S. RICHARDS, G. M. WEINSTOCK, R. A. GIBBS et al., 2006. Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of Holometabolous insects. Genome Research 16: 1334-1338. 17. WOLF, Y. I., I. B. ROGOZIN and E. V. KOONIN, 2004. Coelomata and not Ecdysozoa: evidence from genome-wide phylogenetic analysis. Genome Research, 14: 29-36. 18. KEELING, P. J., G. BURGER, D. G. DURNFORD, B. F. LANG, R. W. LEE et al., 2005. The tree of eukaryotes. Trends Ecology and Evolution, 20: 670-676. 19. PURVIS, A., and A. RAMBAUT, 1995 Comparative analysis by independent contrasts (CAIC): an Apple Macintosh application for analysing comparative data. Computer Application in Biosciences, 11: 247-251. 20. GRAFEN, A.1989. The phylogenetic regression. Philosophical Transactions of the Royal Society. London B. 326: 119–156.