Size of each fragment in base pairs
advertisement

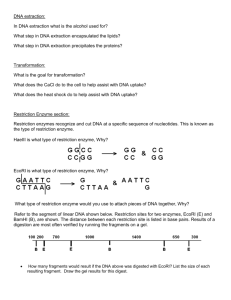
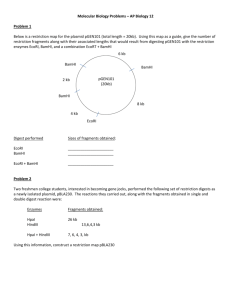
DNA Scissors An Introduction to Fragment Cutting in Plasmid and Linear forms of DNA. The picture below is a restriction map of a plasmid. Restriction sites, where enzymes cut at specific base pairs, are shown on the map. The numbers after the enzyme names tell at what base pair the enzyme cleaves the DNA. There is an EcoRI site at base pair 1. Your job is to determine the length of the DNA fragments after the enzyme or enzymes are used to make their cuts. 1. How long is the entire plasmid?__________________________________ 2. Complete the following table for each restriction digest: Restriction Digest of Plasmid with: EcoRI # of fragments Size of each fragment in base pairs BamHI and HindIII EcoRI,BAMHI and HindIII 10,000 base pairs total BamHl 7,000 EcoRl 1/10,000 PlaPl EcoRl 5,000 Hindlll 2,600 3. What is the relationship between the number of cut sites and the number of DNA fragments for plasmid DNA? The figure below is the map of the restriction enzyme sites of a plasmid named YIP5. This plasmid contains 5,541 base pairs. The number after each enzyme name tells where the enzyme cuts on the plasmid. HindIII 32 EcoRI 1/5,541 PvuI 4916 EagI 942 YIP5 ApaI 2036 PvuII 3247 SmaI 2540 4. Complete the chart for each of the restriction digests listed below. Remember, to check your work by making sure that the sum of the fragments is 5,541 for each question. Restriction Digest of Plasmid with: EcoRl EcoRl and Eagl Hindlll and Apal EcoRl, Eagl, and Pvull Hindlll, Apal, and Pvul Smal, Apal, and Pvull # of fragments Size of each fragment in base pairs Linear DNA cuts can be determined in a similar way. Below are 3 linear pieces of the same DNA. It is 5,000 base pairs long. Map the following cuts and complete the table below. Restriction Digest of Linear DNA with: EcoRl cuts at 1,500 and 3,500 #of fragments Size of each fragment in base pairs BamHl cuts at 2300, 3,900 and 4,500 Both EcoRl and BamHl 1 5000 1 5000 1 5000 EcoRl BamHl EcoRl & BamHl What is the relationship between the number of cut sites and the number of DNA fragments for linear DNA? The plasmid below has the restriction sites as listed. Total length of the plasmid is 20,000 base pairs. Draw the fragments for each digestion as they would appear after electrophoresis. Below each line write the number of base pairs. No enzymes EcoRI HindIII BamHI EcoRI & BamHI Sample Wells HindIII 17,000 BamHI 1/20,000 Size Scale in Base pairs 8000 6000 4000 EcoRI 14,500 3000 BamHI 5,500 EcoRI 12,000 2000 EcoRI 9,000 1000 500 Below are four pieces of the same 15,000 base pair DNA molecule. Use a vertical line to show the cut sites for the digestion with EcoRI & BamHI. EcoRI Draw the fragments for each digestion as they would appear after electrophoresis. Below each line write the number of base pairs. No enzymes 4000 3500 2500 5,000 Sample Wells Size Scale in Base pairs 8000 BamHI 6000 6000 4000 3000 2000 4000 HindIII 3000 8000 4500 2500 2000 EcoRI and BamHI 1000 500 EcoRI HindIII BamHI EcoRI & BamHI