Distinguishing Bacteria by 16S ITS Sequencing
advertisement

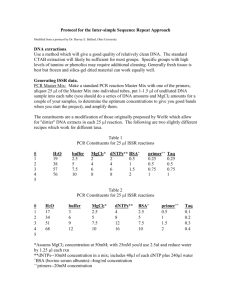
Distinguishing Bacteria by 16S ITS Sequencing Purpose Identification of bacteria by direct sequencing of 16S regions. Materials ABI GeneAmp 9700 OmniMix HS master mix beads (primary method, Cepheid) or 50x Titanium Taq DNA polymerase (secondary method, Clontech) Primers, primary method (Weisburg et al, 1991): Forward primer fD1 (AGAGTTTGATCCTGGCTCAG, most eubacteria,) Reverse primer rP1 (ACGGTTACCTTGTTACGACTT, enterics and most eubacteria) Primers, secondary method: Forward Primer U3 (AGTGCCAGCAGCCGCGGTAA, James, G. 2010) Reverse Primer U4 (AGGCCCGGGAACGTATTCAC, James, G. 2010) Forward Primer U515 (TGCCAGCAGCCGCGGTAATAC, Han, X. Y. et. al., 2002) Reverse Primer U1087 (CGCTCGTTGCGGGACTTAACC, Han, X. Y., et. al. 2002) Instructions Sample preparation: Prepare an unquantified translucent suspension of a pure 3-day culture. Primer preparation: Make 5 µM suspensions in 1x TNE buffer. Primary Method: Using one OmniMix HS bead per 50 µl rxn, add primers fD1 and rP1 to a final 250 nM concentration and add sterile molecular-grade water for a 49 µl reaction volume, then add 1 µl dilute suspended bacterial cells to the reaction. Protocol name, primary method: Weisburg16s. rdna Thermocycling settings, primary method: Initial hold at 95°C for 120 seconds 35 cycles of 95°C for 20 seconds, 55°C for 20 seconds, and 72°C for 20 seconds 8 to 20 minute extension step at 72°C. Note: In our trials, most of our samples amplified well using an eight-minute final extension; however, one bacterial amplicon did not amplify as efficiently as the others and would probably benefit from the additional final extension time. Secondary method: Protocol name, secondary method: 16s-rdna Thermocycling settings, secondary method: Initial hold at 95°C for 120 seconds 30 cycles of 96°C for 15 seconds, 60°C for 90 seconds, and 72°C for 120 seconds Final 72°C extension for 300 seconds This is a 2.5 hour protocol. Primer pairs used: U3 with U4 U515 with U1087. Using one OmniMix HS bead per 50 µl rxn, add primers to a final 250 nM concentration each, add sterile molecular-grade water for a 49 µl reaction volume, then add 1 µl dilute suspended bacterial cells to the reaction and amplify using the above thermocycling settings. Expected results Primary method: Amplicon size is determined by running an agarose gel, and is expected to be approximately 1500 base pairs. Secondary method: Amplicon size is determined by running an agarose gel, and is expected to be approximately 650 bp using primers U3 and U4 (this CAN yield an additional larger band with some isolates, in addition to the primary band. The primer pair U515 and U1087 yields a band of approximately 550 bp and no apparent secondary bands. Sequencing of amplicons Run a 5 µl volume on an agarose gel to verify amplification and product size. Run the remaining 40 – 45 µl on a 1.5% gel and then excise the band. Clean it using QiaQuick gel extraction kit (Qiagen, cat. no. 28704), taking care to follow instructions for direct sequencing. Elute in 30 µl Buffer EB. For amplicons generated using primers U3 and U4, sequence with U4 only (cost savings, better sequence quality with this primer) For amplicons generated using primers U1087 and U515, sequence with U1087 only (cost savings) Notes Using the secondary methods, somewhat successful, but less so in our hands, a pair of primers (U1 with U2) are also commonly used (James, G., 2010). Resources Han, X. Y., MD, PhD, Audrey S. Pham, PhD, Jeffery J. Tarrand, MD, Pramila K. Sood, MBA, and Rajyalakshmi Luthra, PhD. 2002. Am. J. Clin. Pathol. 118:796-801. James, G. 2010. Universal Bacterial Identification by PCR and DNA Sequencing of 16S rRNA Gene. PCR for Clinical Microbiology. M Schuller et al. (eds.) DOI 10.10007/978-90-4819039-3_28, ©Springer Science+Business Media B.V. Weisburg, William G., Susan M. Barns, Dale A. Pelletier, and David J. Lane. 1991. J. Bacteriol. Jan. pp. 697-703.