mec13374-sup-0008-AppendixS2
advertisement

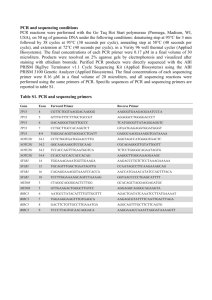
Appendix S2: Molecular Methods Mitochondrial amplification The complete mitochondrial cytochrome-b gene (1140 base pairs) was amplified using universal primers L14723 and H15915 (Table 1, Irwin et al. 1991). Total reaction volume of 25 µl included 100-200 ng of DNA, 0.12 µM of each primer, 1.5 mM MgCl2, 0.012 mM each dNTPs, 1X reaction buffer, 0.32 mg/mL Bovine Serum Albumin and 0.625 U GoTaq Taq polymerase (Promega Corporation, Madison, Wisconsin, USA). The thermal profile included an initial denaturing step at 95oC for 3 min followed by 34 cycles of 95oC for 1 min, 50oC for 1 min and 72oC for 1 min, followed by a final extension time of 72oC for 10 min. Nuclear amplification for X- and Y-chromosome markers Amplicons of approximately 800 bps in length of intron eight of transketolase-like 1 (TKTL1) gene on the X-chromosome were amplified using unpublished primers (Table 1). Amplicons of approximately 940 bps in length of intron 11 of Ubiquitously Transcribed Tetratricopeptide Repeat Containing gene (UTY) were amplified using primers outlined in Hellborg and Ellegren (2003) (Table 1). The total reaction volume of 25 µl included 100-200 ng of DNA, 0.5 µM of each primer, 0.125 mM each dNTPs, 1X reaction buffer, 0.15 X DMSO and 1 U Phire Hot Start II DNA Polymerase (Thermo Fisher Scientific, Pittsburg, PA, USA). The thermal profile included an initial denaturing step at 98oC for 1 min 45 sec followed by 34 cycles of 98oC for 20 sec, anneal for 20 sec and 72oC for 40 sec. The annealing temperatures selected for each primer combination were two degrees higher than that estimated for the primer with the lower melting point (Table 1). Sanger sequencing Cycle sequencing was performed with BigDye version 3.1. (Applied Biosystems, Inc., Foster City, California, USA) and primers listed in Table 1 with reaction conditions following manufacturers protocols. Sequences were electrophoresed on and ABI 3130 (Applied Biosystems, Inc., Foster City, California, USA) sequencing platform. AFLP methods The AFLP protocol followed Vos et al. (1995) with thermal profile modifications described by McDonough et al. (2008). Genomic DNA was digested with AseI and EcoRI restriction enzymes (New England Biolabs Inc., Ipswich, Massachusetts, USA). Preselective PCR was performed using primers AseI-T and EcoRI-C and selective PCR was performed using seven primer pairs on all individuals (Table 2). Selective PCR products along with Genescan-400HD ROX size standard were electrophoresed on an ABI 3100-Avant genetic analyzer (Applied Biosystems, Inc., Foster City, California, USA). Fragments ranging from 50 to 400 nucleotides were scored for presence or absence of bands using GeneMapper version 4.0 software (Applied Biosystems). All ambiguous fragments (i.e., fragments that were weakly amplified in some individuals) were removed from the final data set. Error rate and reproducibility were evaluated by replicating 10% of the reactions as suggested by Bonin et al. (2004) and an error was considered as an amplification that was inconsistent between replicates. Table 1. Primers used for PCR and sequencing. Primer Name CYTB L14723 H15915R UTY UTY11F UTY11R TKTL1 PCR TKTL1_X8Fb TKTL1_X9Rb Sequencing TKTL1_X8Fc TKTL1_X9Rc Primer sequence Tm (DegC) 5'-ACCAATGACATGAAAAATCATCGTT-3' 5'-GGAATTCATCTCTCCGGTTTACAAGA-3' 61.4 63.4 5'-CATCAATTTTGTAYMAATCCAAAA-3' 5'-TGGTAGAGAAAAGTCCAAGA-3' 53.2 50.9 5'-GCCATCTCTCAAATCAACAT-3' 5'-GGCTGCTAAGTAAACAGCAT-3' 53.5 53.4 5'-YGATTGCTGGAAACAGGAAA-3' 5'-CCCAAGGCTTTGTCTTTCA-3' Table 2. Primers used for selective amplification of amplified fragment length polymorphism (AFLP) bands. Asterisk indicates fluorescently labeled primer. Primer Name EcoRI-CAT* AseI-TAC AseI-TAG AseI-TCT AseI-TGC AseI-TGT AseI-TGA AseI-TGG Sequence 5'-GACTGCGTACCAATTCCAT-3' 5'-GATGAGTCCTGAGTAATTAC-3' 5'-GATGAGTCCTGAGTAATTAG-3' 5'-GATGAGTCCTGAGTAATTCT-3' 5'-GATGAGTCCTGAGTAATTGC-3' 5'-GATGAGTCCTGAGTAATTGT-3' 5'-GATGAGTCCTGAGTAATTGA-3' 5'-GATGAGTCCTGAGTAATTGG-3' References Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetics studies. Molecular Ecology, 13, 3261–3273. Hellborg L, Ellegren H (2003) Y chromosome conserved anchored tagged sequences (YCATS) for the analysis of mammalian male-specific DNA. Molecular Ecology, 12, 283-291. Irwin DM, Kocher TD, Wilson AC (1991) Evolution of the Cytochrome b gene of mammals. Journal of Molecular Evolution, 32, 128-144. McDonough MM, Ammerman LK, Timm RM, Genoways HH, Larsen PA, Baker RJ (2008) Speciation within Bonneted bats (genus Eumops): the complexity of morphological, mitochondrial, and nuclear data sets in systematics. Journal of Mammalogy, 89, 1306– 1315. Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research, 23, 4407–4414.