MicrobialDiversity_Session2
advertisement

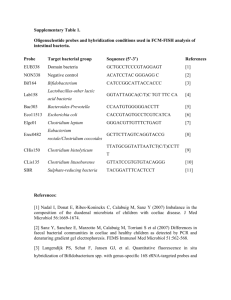
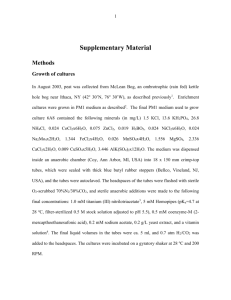
David Zepeda-Campos Katia Garcia Hector Ibarra Ashleigh Richelle Topics in Science: Presentation #2 Microbial Diversity Have you ever wondered what kinds of things live in your refrigerator or bathroom? What about in creeks or the inside of your mouth? In our group, we were able to answer some of those questions. During our group sessions, we collected samples from these places, incubated them, prepared them and viewed them under the light microscope, and finally we used the Fluorescence In Situ Hybridization method (FISH), to identify the organisms present. The key microorganisms we viewed under the microscope with and without the FISH method were fungi and bacteria. We were able to isolate these microorganisms from the sample we had collected, and the outcome of the work we had done was really interesting and amazing to look at. Our first day, we were each handed a Petri dish plate with nutrient rich-medium in it and were told that our task was to find a place where we could leave it open or swab in order to find out what kind of microorganisms grew there. We each chose different places. David and Hector decided to leave it in a refrigerator, Katia left hers in the girls’ bathroom while Ashleigh used swabs from dirt water, her mouth and a garbage can. After completing that task, we incubated the plates at 37C for 2 days. On Tuesday, we found that we are surrounded with more bacteria than we thought. Ashleigh even got some fungi to grow in hers. Once we had observed them with our naked eye, we used a dissection microscope. This allowed us to see colony morphology and see variety of shapes and colors, such as blob-shaped colonies to circular-shaped colonies, as well as colors like whites, yellows and browns. Another characteristic that was evident our first day investigating was the horrible stench being emitted by the various bacteria. We were also able to identify hyphae which are the filaments from fungi. To see the cell morphology we each took our sample, placed it on a slide and used the light microscope. This microscope was connected to a computer. It was able to take pictures and project a bigger image on the computer screen. When we were looking at the image projected by the microscope on the computer, we were able to see that the bacteria were still alive! Some of them were moving across and others were just moving in place. At this point we were able to categorize the bacteria by their shape including circular-shaped bacteria called cocci and rod-shaped bacteria called bacilli The FISH protocol is an essential method in microbiology for narrowing down and identifying microorganisms that may be present in an environment setting. The base of this method all starts off with a molecule called 16S rRNA. The ribosomal RNA (rRNA) is present in all organisms and it is involved in the synthesis of proteins. This specific type of ribosomal RNA is given its name (16S) due to its density after centrifugation. Such variety of the 16S rRNA in organisms is due to the evolution and natural selection of the organisms. While all organisms contain this rRNA, there are a variety of sequences among the population. This procedure, however, is not limited to prokaryotic cells. It can also be used for eukaryotic cells; the difference though is that the probe used to identify the eukaryotic cells would be targeting 23S rRNA. This method works by designing 16S rRNA probes and adding them to a sample of bacteria. These probes are designed to target a specific sequence in a prokaryotic cell and “hybridize” or fuse to it. Probes can be designed to identify short regions in a sequence ranging from twelve to twenty-five nucleotides in length. Once the protocol is done, scientists can assess such characteristics like phylogenetic identity, morphology, number, and spatial arrangements. For the FISH process, we first had to fix the sample on a slide. We used a specific kind of slide which was actually separated into eight sections so we would have enough room to place all our samples together, but apart from each other at the same time. The samples that we each used were from the colonies of bacteria that we obtained from the different locations we chose. We scooped up small portions of the various colonies with a loop tool, placed the samples in eppendorf tubes and added 500ul of fixative (PFA 4%). After incubating the samples for about two hours at four degrees C, we centrifuged the tubes, and added one volume of PBS ph 7.0, plus one volume of 100% ethanol. After we had fixated the solution, we went into the actual F.I.S.H. procedure. The first things we had to do were turn on a Bunsen burner, so to kill off any and all unnecessary bacteria floating in the air. Then we put on our gloves. After putting 5ul of each sample on four of the wells (one per person) of the FISH slide and letting it air dry, we had to make the hybridization buffer in a 2ml eppendorf tube. We did this by adding 898ul of miliQ water, 700ul of formamide, 360ul of NaCl 5m, 40ul of buffer Tris-HCl ph 8.0, and 2ul of 10% SDS. We then dehydrated the slide in 50%, 80%, and 90% ethanol solution for three minutes each. After letting it dry again from the dehydration process, we prepared the hybridization chamber by placing a kimwipe in a 50ml tube and adding 1.8ul of hybridization buffer followed by 8ul of the buffer to each well and 1ul of each probe as well. We then placed the slide in the hybridization chamber and incubated it again at 46 degrees C for 2 hours. After that, we rinsed the slide with washing buffer which consisted of the same thing the hybridization buffer has. We rinsed the slide one last time with the washing buffer in order to get rid of any leftover probes. After the slide dried, we saw it through a light microscope. The FISH protocol detects the different organisms by lighting them up a different color. There are 4 probes that hybridize, or attach, to the bacteria and make them light up. The probe for eukaryotes is green, the probe for general bacteria is blue, the probe for Gama probacteria is green, and the probe for firmicutes is red. Sometimes some microorganisms will hybridize with two probes, so there will be more combinations of colors that can result from this process. If an organism attaches to a blue and a green probe, then the organism will show as light blue. If an organism attaches to a red and a green probe, then it will show up as yellow. If an organism attaches to a blue and a red probe, then it will show up as purple. And finally, if an organism attaches to a red, blue, and green, then it will show up as white. Our results ultimately showed that we had almost every kind of the organisms we went looking for. Practically every color on the chart was evident in our visualization of the bacteria samples. From collecting the samples to preparing them and viewing them under the light microscope, the overall experience of finding out was a great one. We were able to see that sometimes the places we would love to keep really clean, like our mouths and refrigerators, can’t always be kept clean. There will always be something living in or around us other than humans and that is what makes life interesting and diverse in many ways.