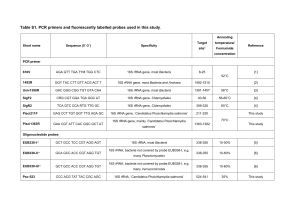
Target Primer Sequence Region Reference Bacteria B8F
advertisement

Target Primer Bacteria Bacteria Universal Bacteria Bacteria Archaea Bacteria Universal Universal Bacteria Sequence Region B8F AGAGTTTGATCCTGGCTCAG B533R TTACCGCGGCTGCTGGCAC U341F CCTACGGGRSGCAGCAG B785R TACNVGGGTATCTAATCC U341F CCTAYGGGRBGCASCAG U806R GGACTACNNGGGTATCTAAT U341F CCTAYGGGRBGCASCAG B907R CCGTCAATTCMTTTGAGTTT B343F TACGGRAGGCAGCAG B534R ATTACCGCGGCTGCTGGC A344F ACGGGGYGCAGCAGGCGCGA A915R GTGCTCCCCCGCCAATTCCT U347F GGAGGCAGCAGTRRGGAAT U803R CTACCRGGGTATCTAATCC U515F GTGYCAGCMGCCGCGGTA U806R GGACTACHVGGGTWTCTAAT U515F GTGYCAGCMGCCGCGGTA U909R CCCCGYCAATTCMTTTRAGT U519F CAGCMGCCGCGGTAATWC B926R CCGTCAATTCMTTTRAGTT V1-V3 Reference (Wang and Qian, 2009) V3-V4 (Klindworth et al., 2013) V3-V4 (Zakrzewski et al., 2012) V3-V5 (Wang and Qian, 2009; Sogin et al., 2 006) V3 (Liu et al., 2007) V3-V5 (Teske and Sorensen, 2008) V3-V4 (Nossa et al., 2010) V4-V5 (Caporaso et al., 2011) V4-V5 (Tamaki et al., 2011) V4-V5 (Mao et al., 2011) References 1. Caporaso, J.G., Lauber, C.L., Walters, W.A., Berg-Lyons, D., Lozupone, C.A., Turnbaugh, P.J., Fierer, N., Knight, R., 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America 108 Suppl 1, 4516-4522. 2. Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., Glockner, F.O., 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research 41,e1. 3. Liu, Z., Lozupone, C., Hamady, M., Bushman, F.D., Knight, R., 2007. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Research 35, e120. 4. Mao, Y., Yannarell, A.C., Mackie, R.I., 2011. Changes in N-transforming archaea and bacteria in soil during the establishment of bioenergy crops. PloS One 6, e24750. 5. Nossa, C.W., Oberdorf, W.E., Yang, L., Aas, J.A., Paster, B.J., Desantis, T.Z., Brodie, E.L., Malamud, D., Poles, M.A., Pei, Z., 2010. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World Journal of Gastroenterology 16, 4135-4144. 6. Sogin, M.L., Morrison, H.G., Huber, J.A., Mark Welch, D., Huse, S.M., Neal, P.R., Arrieta, J.M., Herndl, G.J., 2006. Microbial diversity in the deep sea and the underexplored "rare biosphere". Proceedings of the National Academy of Sciences of the United States of America 103, 12115-12120. 7. Tamaki, H., Wright, C.L., Li, X., Lin, Q., Hwang, C., Wang, S., Thimmapuram, J., Kamagata, Y., Liu, W.T., 2011. Analysis of 16S rRNA amplicon sequencing options on the Roche/454 next-generation titanium sequencing platform. PloS One 6, e25263. 8. Teske, A., Sorensen, K.B., 2008. Uncultured archaea in deep marine subsurface sediments: have we caught them all? The ISME journal 2, 3-18. 9. Wang, Y., Qian, P.Y., 2009. Conservative Fragments in Bacterial 16S rRNA Genes and Primer Design for 16S Ribosomal DNA Amplicons in Metagenomic Studies. PLoS One 4(10):e7401. 10. Zakrzewski, M., Goesmann, A., Jaenicke, S., Junemann, S., Eikmeyer, F., Szczepanowski, R., Al-Soud, W.A., Sorensen, S., Puhler, A., Schluter, A., 2012. Profiling of the metabolically active community from a production-scale biogas plant by means of high-throughput metatranscriptome sequencing. Journal of Biotechnology 158, 248-258.