Growth of cultures
advertisement

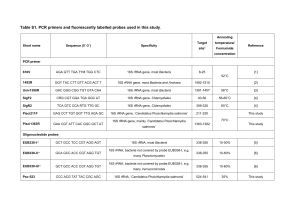
1 Supplementary Material Methods Growth of cultures In August 2003, peat was collected from McLean Bog, an ombrotrophic (rain fed) kettle hole bog near Ithaca, NY (42o 30’N, 76o 30’W), as described previously1. Enrichment cultures were grown in PM1 medium as described2. The final PM1 medium used to grow culture 6A8 contained the following minerals (in mg/L) 1.5 KCl, 13.6 KH2PO4, 26.8 NH4Cl, 0.024 CoCl2x6H2O, 0.075 ZnCl2, 0.019 H3BO3, 0.024 NiCl2x6H2O, 0.024 Na2Mo4x2H2O, 1.344 FeCl2x4H2O, 0.026 MnSO4x4H2O, 1.556 MgSO4, 2.336 CaCl2x2H2O, 0.009 CuSO4x5H2O, 3.446 AlK(SO4)2x12H2O. The medium was dispensed inside an anaerobic chamber (Coy, Ann Arbor, MI, USA) into 18 x 150 mm crimp-top tubes, which were sealed with thick blue butyl rubber stoppers (Bellco, Vineland, NJ, USA), and the tubes were autoclaved. The headspaces of the tubes were flushed with sterile O2-scrubbed 70%N2/30%CO2, and sterile anaerobic additions were made to the following final concentrations: 1.0 mM titanium (III) nitrilotriacetate3, 5 mM Homopipes (pKa=4.7 at 28 oC, filter-sterilized 0.5 M stock solution adjusted to pH 5.5), 0.5 mM coenzyme-M (2mercapthoethanesufonic acid), 0.2 mM sodium acetate, 0.2 g/L yeast extract, and a vitamin solution4. The final liquid volumes in the tubes were ca. 5 ml, and 0.7 atm H2/CO2 was added to the headspaces. The cultures were incubated on a gyratory shaker at 28 oC and 200 RPM. 2 For experiments on the effect of pH on culture 6A8, Homopipes was omitted and the pH of the medium was adjusted by addition of 10 mM citrate adjusted to various pH values. The pH of the cultures was assessed at the end of incubations. For experiments on the effect of temperature, cultures were incubated under static conditions. Methane production by cultures was quantified using a flame ionization gas detector as previously described1. Molecular and phylogenetic analyses A MoBio Ultraclean Microbial DNA extraction kit that includes a bead-beating step was used to extract and purify DNA from the culture. Primers 1Af(-1) (TCYGKTTGA TCCYGSCRGA) (a 3'-G was deleted from the original primer5 to increase range) and 1100Ar (TGGGTCTCGCTCGTT G) were used to amplify methanoarchaeal 16S rRNA genes and 27f (AGAGTTTGATCMTGGCTCAG) and 1492r (TACGGYTACCTTGTTACGACTT) were used to amplify bacterial 16S rRNA genes. The PCR reaction contained 1.25 units Brinkmann Taq polymerase, 1x Brinkmann Taq buffer, 0.18 µM primer, 200 µM dNTPs, and 4 µl of a 1:1 diluted DNA extract in a total volume of 55 µl. For both the bacterial and archaeal amplifications, the program consisted of 4 minutes at 95 oC, then 25 cycles of 95 oC for 1 minute, 50 oC for 1 minute, and 72 oC for 1.5 minutes, then a final elongation step at 72 oC for 10 minutes. To obtain an amplicon containing the 16S rRNA gene, the internal transcribed spacer region (ITS, ca. 200 bases), and ca. 50 bases of the 23S rRNA gene, the primers 1AF (TCYGKTTGATCCYGSCRGAG)5 and ArchLSU47 (CTTATCGCAGCTTRSCACG) were used, producing an amplicon of 1730 bases. The PCR program was 4 minutes at 95 oC 3 followed by 25 cycles of 94 oC for 0.5 minutes, 53 oC for 1 minute, and 72 oC for 2 minutes, followed by a final incubation at 72 oC for 6 minutes. The PCR products were screened on a 1% agarose gel in TBE buffer. Ligations and transformations were performed using the Invitrogen TA Cloning kit. Potential transformants were screened using the M13f(-20) and M13r(-27) primers using the following amplification program: 4 minutes at 95 oC, then 35 cycles of 95 oC for 1 minute, 46 oC for 1 minute, and 72 oC for 1.5 minute, then an elongation step of 72 oC for 10 minutes. Clones with proper sized inserts were digested overnight using restriction endonucleases HaeIII and HhaI6 (New England BioLabs). Representatives of restriction digestion types were sent to Cornell's Bioresource Center for sequencing. Most sequences used for phylogenetic analyses of 16S rRNA genes were either prealigned in the 2003 ARB database7 or were aligned using the ARB sequence editor, except for AMC1, KB-1, FenH1 and K4-a2, which were added using CLUSTALX to an alignment exported from ARB. The tree presented in Fig. 2 was produced from 956 bases (many of the environmental sequences were <1100 bases) using the DNAML maxiumum likelihood program with the global rearrangement option from the PHYLIP 3.62 package8. Bootstrap values were using SEQBOOT, DNADIST using the F84 distance model, and NEIGHBOR from PHYLIP. Fluorescence microscopy Acridine orange (AO) staining was performed by adding 10 µL of cell culture and 1 µl 0.01% w/v AO for each wet mount. For examination of cells by fluorescence in situ hybridization (FISH), cells were fixed in 4% paraformaldehyde for 15-24 hours, filtered 4 onto black 0.2-µM polycarbonate membrane filters, and stored with desiccant at –20 oC. FISH was performed as described9 with some modifications. Hybridization buffer contained 0% formamide and 2 µg/mL DAPI (4’, 6-diamidino-2-phenylindole) and one of the following indocarbocyanine dye-labeled (Cy-3-labeled) probes at a final concentration of 2 ng/µL (EUB 33810, NON EUB 338 5’Cy-3 – ACTCCTACGGGAGGCACG, ARCH 91511, MG120011, MG12006A, 5’Cy-3 – CGGACAATTCGGGGCATGCTG, or 6A8 644 5’Cy-3 – TCTTCCGGTCCCTAGCCTGCCA (E. coli numbering 644-665)). Oligonucleotide probe 6A8 644 targeting small subunit rRNA from methanogen 6A8 was designed using the probe design and probe match tools in ARB software7 using the August 2003 database, to which we added over 200 archaeal sequences including many from our studies. This probe had five mismatches to Methanospirillum hungatei, the closest cultured relative to 6A8, and of the sequences in Fig. 2, only matched KB-1 and R10 perfectly, with a single mismatch to other members of the E2 group and two mismatches to members of the E1 group. Quartered membrane sections were fixed to slides using tough-tag centrifuge labels and 50 µL of hybridization buffer were added to each membrane section, which was then covered with square 22 mm cover slips. Reactions were conducted in a high humidity chamber at 37 oC overnight. Hybridization stringency was optimized according to Morris et al.12 by washing at a temperature near the empirically determined disassociation temperature for each probe. Membrane sections were washed in two 10-minute intervals in wash solution (.2X SET 9, without formamide). Membrane sections were dried, mounted with Citifluor and a cover slip, and viewed using a Nikon Eclipse E600 epifluorescence microscope equipped with a Hamamatsu CCD digital camera, or using an Olympus BX61 epifluorescence microscope equipped with a Cooke SensiCam camera with a Sony Interline 5 Chip and the SlideBook Software package (Intelligent Imaging). Consistent exposure times (3.0 seconds), histogram thresholds and contrast values were used for all cy3 images made with the species specific probe 6A8 644, with positive controls made with the archaeal probe ARCH 915 and with negative controls made with the NON EUB 338 probe. Exposure times for cy3 images made with probe MG12006A8 were auto-calibrated by the slide book software and were approximately three seconds. Exposure times for the DAPI images were all approximately one second and were auto-calibrated by the SlideBook software. References 1. Bräuer, S. L., Yavitt, J. B. & Zinder, S. H. Methanogenesis in McLean Bog, an acidic peat bog in upstate New York: Stimulation by H2/CO2 in the presence of rifampicin, or by low concentrations of acetate. Geomicrobiol J 21, 433-443 (2004). 2. Bräuer, S. L., Yashiro, E., Ueno, N. G., Yavitt, J. B. & Zinder, S. H. Characterization of acid-tolerant H2/CO2-utilizing methanogenic enrichment cultures from an acidic peat bog in New York State. (in press). 3. Moench, T. T. & Zeikus, J. G. An improved preparation method for a titanium(III) media reductant. J Microbiol Methods 1, 199-202 (1983). 4. Balch, W. E., Fox, G. E., Magrum, L. J., Woese, C. R. & Wolfe, R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev 43, 260-296 (1979). 6 5. Hales, B. A. et al. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl Environ Microbiol 62, 668-75 (1996). 6. Moyer, C. L., Tiedje, J. M., Dobbs, F. C. & Karl, D. M. A computer-stimulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrametric restriction enzymes for studies of microbial diversity in nature. Appl Environ Microbiol 62., 251-257 (1996). 7. Ludwig, W. et al. ARB: a software environment for sequence data. Nuc Acids Res 32, 1363-1371 (2004). 8. Felsenstein, J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle. (2004). 9. DeLong, E. F., Taylor, L. T., Marsh, T. L. & Preston, C. M. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol 65, 5554-5563 (1999). 10. Amann, R. I. et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56, 1919-25 (1990). 11. Raskin, L., Stromley, J. M., Rittmann, B. E. & Stahl, D. A. Group specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol 60, 1232-1240 (1994). 7 12. Morris, R. M. et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420, 806-810 (2002). 13. Boone, D. R. & Castenholz, R. W. Bergey's manual of systematic bacteriology. 2nd Ed. Vol 1. The Archaea and deeply brainching and phototrophic Bacteria. (ed. Garrity, G. M.) (Springer, New York, 2001). 14. Basiliko, N., Yavitt, J. B., Dees, P. M. & Merkel, S. M. Methane biogeochemistry and methanogen communities in two northern peatland ecosystems, New York State. Geomicrobiol J 20, 563-577 (2003). 8 Supplementary Figures Supplementary Figure 1. Phylogenetic tree for 16S rRNA gene sequences from culture 6A8, selected sequences in the Methanomicrobiales showing presently accepted families 13, and other members of the Euryarchaeota. Sequences labelled PBC are peat bog clones, those labelled MB were from a McLean Bog clone library14, and enrichment culture clones are labelled ECC. The tree was constructed by the maximum likelihood method (see methods) and nodes with bootstrap values > 80% (100 replicates) from a neighbour-joining method are labelled. Methanomicrobiales Sulfolob us acidocaldariusU05018 Methanococcus maripaludisAF005049 M etha nococca les 100 MethanocaldococcusjannaschiiU67473 M etha nopyra les Methanopyrus kandleriM59932 Methanothermus fervidus M59145 M etha noba cteria les 99 Methanob acterium b ryantiiM59124 Archa ea ogloba les Archeoglob us fulgidusAE000965 Methanosarcina acetivoransM59137 100 M etha nosa rcina les Methanosaeta conciliiM59146 95 Methanoculleus b ourgensisAF095269 100 Methanomicrob ium mob ileAF095269 Methanomicrob iaceae Methanogenium cariaci M59130 Methanocorpusculaceae Methanospirillum hungateiM60880 100 Methanocorpusculumparvum M59147 Methanospirillaceae PBC MB-10 AY175398 100 E1 PBC MB-06 AY175393 PBC MB-16 AY175403 PBC MB-03 AY175390 0.1 R10 group, 99 ECC KB-1 1 AY780566 Fen cluster, 86 Strain6A8 DQ282124 E2 E1/E2 group ECC K4-a2 AF524852 ECC AMC1 AJ459899 90 PBC R10 L48407 PBC FenH1 AJ548948 9 Supplementary Figure 2. Fluorescence micrographs of 6A8 stained with DAPI or with a Cy-3-labelled 16S rRNA probe MG12006A8 (a,b) or Methanospirillum hungatei JF1 cells stained with the DNA stain DAPI (c,e) or with a Cy-3-labelled16S rRNA probe targeting Archaea, probe Arch 915 (d), or targeting 6A8, probe 6A8 644(f). (a) (b) (c) (d) (e) (f) 10 Supplementary Figure 3. Effect of temperature on methanogenesis by culture 6A8. Data points represent the averages and error bars the standard errors for triplicate samples. Methane Production (mmoles/L) 8 7 Day 3 6 Day 6 Day 24 5 4 3 2 1 0 0 10 20 30 40 o Temperature ( C) 50