restriction digest
advertisement

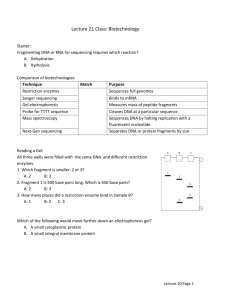
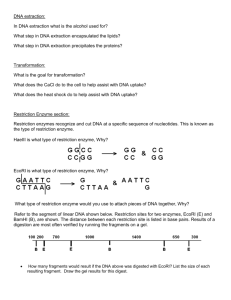
Restriction Digest Molecular Biology Lab #3 Background: Restriction endonucleases are enzymes that have been isolated from bacteria. A large number of these enzymes (over 3300 currently described) have been obtained from a wide variety of bacterial types. Restriction endonucleases cleave within the DNA molecule and at characteristic restriction sites for each specific enzyme. The restriction site is usually composed of 4 to 8 base pairs that are organized in a specific sequence that is recognized by the enzyme. Different restriction enzymes cleave in different patterns to generate blunt or overhanging ends (sticky ends) of the DNA molecule. Many restriction sites are composed of short inverted repeats. Restriction enzymes are an extremely important tool in molecular biology. The restriction enzyme sites within any DNA molecule are readily predicted from the sequence of bases. Restriction digestion is used to separate fragments of genes for manipulation in the lab (i.e. cloning or mutagenesis). Digestion with restriction endonucleases produces a specific pattern of DNA bands when the reaction products are separated by gel electrophoresis. This is termed the restriction pattern. The restriction pattern is diagnostic of a specific DNA sequence, and it is frequently used to confirm the identity of specific DNA fragments (for example, your unknown DNA). Restriction endonucleases serve an important function in bacterial defense. Because they are able to cleave any foreign DNA that enters the bacteria, they are capable of inactivating intracellular bacterial pathogens. Interestingly, the bacteria also contain specific methylases, enzymes that add a methyl group to the restriction site of enzymes that the bacteria produce. This methylation prevents the restriction endonuclease from digesting the bacterial DNA and killing the host cell. The REBASE website http://rebase.neb.com/rebase/rebase.html is a very informative site that contains everything that you would want to know about any restriction endonuclease. You should navigate this site during incubation of your restriction digest and work on the homework assignment. Where can you get restriction enzymes? The easiest source is to purchase them from a biotechnology company. They range in price from relatively cheap (BamHI = $30/2000 units) to very expensive (RsrII = $820/2000 units). The REBASE website links to many biotechnology websites which makes purchasing or comparative shopping for restriction enzymes easy. How do you store restriction enzymes? They are usually stored dissolved in glycerol so that they may be kept at -20º without actually freezing. You just pull the vial out of the freezer and use it. Be careful about keeping the stock tube of enzyme cold. Place it on ice water whenever you are not using it. This will preserve the enzyme activity. Don’t allow the enzyme to remain at room temperature. How much is enough? Usually 10 units of enzyme per microgram of recombinant DNA is plenty. Digest at 37ºC for 1 to 24 hours. Digest longer if you use less enzyme or if you are digesting cellular DNA (i.e. Southern blotting, later in the course). Do all restriction endonucleases work under the same conditions? No, each enzyme requires very specific conditions to work optimally. Usually, the amount of salt is very important. When you buy an enzyme, a small vial of 10x buffer is usually supplied with the enzyme. See the REBASE web site for an explanation of STAR activity. What happens if you need to digest your DNA with 2 different restriction enzymes? This occurs often when a fragment of DNA is cloned into two different restriction sites (directional cloning). It is possible to do a double digest if the 2 enzymes are able to work in the same buffer. This is easy. You just allow both enzymes to digest the DNA at the same time. Sequential digests must be performed if the 2 enzymes cannot work under the same conditions. Then you must digest with one enzyme, precipitate the DNA, resuspend the DNA in an appropriate buffer, and digest with the second enzyme. What can go wrong? Any contaminants in your DNA, such as agarose or phenol, can inhibit the reaction. If the enzyme is left out at room temp for any length of time, it often can lose activity. Objectives: The purpose of this lab is to introduce you to restriction endonucleases and provide practical experience setting up a restriction digest of plasmid DNA. Materials: 37ºC and 60ºC water baths restriction enzymes BamHI and EcoRI React 3 buffer for the reaction Micro pipetters and yellow tips Eppendorf tubes Deionized water to dilute DNA TE buffer to resuspend precipitated DNA BioRad spectrophotometer Quartz cuvette Microcentrifuge Procedure: 1. If the DNA is stored as a precipitate, you should centrifuge for 5 min in a microfuge to pellet the DNA. Resuspend the pellet in 50 to 200 l of 1x TE buffer. TE buffer is a mixture of Tris buffer at pH 8 and EDTA. DNA is very stable when stored in TE buffer. Mix carefully to ascertain that the DNA is completely dissolved. 2 2. Determine the concentration of DNA. Turn on the spectrophotometer and push the DNA/RNA button and then the dsDNA button. Place 700 ml of 1x TE buffer in the quartz cuvette and gently insert it into the cuvette holder of the spec. Be gentle. The quartz cuvette and the instrument are expensive. Close the cover and push the blank button to zero the instrument. 3. Dilute your DNA samples into 1x TE buffer for analysis. Add 5 l of DNA to 1000 l of 1x TE buffer in an eppendorf tube. Mix by shaking the tube. Pipette the 700 l of DNA solution into the cuvette, insert cuvette into the spectrophotometer, and close the lid. Push the sample button to record the absorbance at 260 nm. 4. Read the concentration of DNA from the dial and multiply by your dilution factor (x200) to calculate the actual concentration of your sample. A solution with a value of 1 (A260 measurement) contains about 50 g of DNA / ml. 5. Touch the 260/280 button to calculate the ratio of absorbance at the 2 wavelengths. This value is usually in the range of 1.8 to 2.0 Very low readings suggest contamination of your DNA with other molecules such as protein or phenol. Record all information. 6. Set up your restriction digest in a total volume of 20 l. Add 2 g of each recombinant DNA to a separate eppendorf tube. Add 2 l of the appropriate restriction enzyme buffer to each tube. Use React3 for BamHI and EcoRI. Add deionized water to each tube to bring the total volume to 19 l. Label eppendorf tubes on the cap. 7. Add enzyme. Remove the tube of restriction enzyme from the ice bath and carefully pipette 1 l into each of your reactions. Use a clean tip each time you pipette from the stock tube of enzyme. This prevents contamination of enzyme with your DNA sample. Keep the samples on ice and gently mix by pipetting up and down with a yellow tip. Spin briefly (10 sec) in an eppendorf tube to collect the reaction at the bottom of the tube. 8. Incubate the restriction digest reaction for 1 hour at 37ºC in a water bath. Place each of your tubes in a small floating rack, close caps securely, and move the rack to the water bath. Check periodically to see that the temperature remains at 37ºC and that the caps of tubes remain closed. 9. After 1 hour, transfer your tubes to the 60ºC water bath to inactivate the enzyme. Spin briefly in a microcentrifuge to collect the reaction in the bottom of the tube. 10. Store at -20ºC until next week. If you had sufficient time, you would proceed to load your samples into a minigel to check the pattern of restriction fragments. 11. Clean up your work area! 3