The University of Michigan Transgenic Animal Model Core Thomas
advertisement

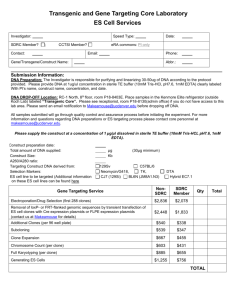
The University of Michigan Transgenic Animal Model Core Thomas L. Saunders, Ph.D. Elizabeth D. Hughes, M.S. Screening Strategy for 2570 MSRB II, SPC 5674 1150 W. Medical Center Drive Ann Arbor, MI 48109-5674 Voice: (734) 647-2910 FAX: (734) 936-2622 http://www.med.umich.edu/tamc/ DATE: Investigator: phone/email: Lab Contact: phone/email: 1. Attach restriction maps of the endogenous gene, targeting vector, and targeted chromosome after homologous recombination. Indicate DNA fragments that will be used as probes on Southern blots, pertinent restriction enzymes, and PCR primers (if applicable). 2. How many Kb of DNA will be deleted from the chromosome by homologous recombination with the targeting vector? 3. Please complete this table: Predicted DNA Fragment Sizes (Kb) on Southern Blot Restriction 5’ Probe Enzyme Endogenous Gene Targeted Gene Random Integrant 3’ Probe Targeted Gene Random Integrant Restriction Enzyme Endogenous Gene 4. Please provide film showing: 5’ Probe - Southern Blot Data on 2-5 µg of ES cell DNA 3’ Probe - Southern Blot Data on 2-5 µg of ES cell DNA Targeting Vector for DATE: Investigator: phone/email: Lab Contact: phone/email: � yes 5. Will a PCR screen be used? � no If so, please provide results that demonstrate the screen is sensitive at the 0.01 genomic copy level (see Cell vol. 62:1073-1085). To achieve this, use a positive control construct which includes sequences recognized by the screening primers. The size of its PCR product should be noticeably different from the size of the product generated by the targeted gene. Information on preparing copy standards in 96-well ES cell DNA is available at http://med.umich.edu/tamc/spike.html. Attach Restriction Map of Vector genetic background of mouse providing arms of homology length of homologous arms: 5’ 3’: were the arms of homology generated by PCR? yes no junction fragments sequenced? � yes � no evidence that 5’ arm is in correct orientation � yes � no evidence that 3’ arm is in correct orientation � yes � no evidence that single bp mutations (if any) are present � yes � no demonstrate screen for single bp mutations (if any) � yes � no demonstrate that loxP sites are in the correct locations (if any) � yes � no demonstrate that loxP sites are in the correct orientation (if any) � yes � no how many base pairs are between the distal loxP site (if any) and the neo cassette? demonstrate that ES cells containing an HSV-TK cassette will not be used to generate chimeras � yes will negative selection be used? � HSV-TK � DT-A If Other, please provide map and description of selection cassette. � no � Other Targeting Vector for DATE: Investigator: phone/email: Lab Contact: phone/email: what is source of neo cassette? � pPNT � pNZ-TK2 � pMC1NEO If Other, please provide map and description of neo cassette source. � Other what restriction site will be used to linearize targeting vector? what size is the targeting vector after linearization? Attach the pertinent paperwork: for R1 ES cells: copy of approved material transfer agreement: � yes � no for CJ7 cells: copy of letter agreeing to acknowledge Tom Gridley: � yes � no for pPNT: copy of letter requesting use of vector: � yes � no for pNZ-TK2: copy of correspondence permitting use of vector: � yes � no Other agreements (DT-A, GFP, IRES, lacZ, etc.)? UCUCA Approval Number The University of Michigan Transgenic Animal Model Core Sally A. Camper, Ph.D. Linda C. Samuelson, Ph.D. Thomas L. Saunders, Ph.D. 2560 MSRB II, Box 0674 1150 W. Medical Center Drive Ann Arbor, MI 48109-0674 Voice: (734) 647-2910 FAX: (734) 936-2622 http://www.med.umich.edu/tamc/ Production of Gene Targeted Embryonic Stem Cell Clones Statement of Understanding of the Nature of the Gene Targeting Service For production of gene targeted embryonic stem cells in collaboration with the Transgenic Core I am responsible for the following tasks: Restriction map the endogenous gene. Establish a robust, reliable, reproducible screen that discriminates between wild type and targeted alleles. Provide experimental data demonstrating that the Southern blot probes will confirm the genomic structure of targeted ES cell clones. Clone the gene targeting vector. Purify gene targeting vector DNA for electroporation according to the protocol supplied by the Transgenic Core. Analyze genomic DNA from as many as 480 ES cell clones for homologous recombination with the targeting vector. Timely analysis is important because of the limited life span of ES cells cryopreserved at 80°C. Review the screening data with the Transgenic Core prior to expansion of cryopreserved clones. Although 480 clones should contain multiple targeted clones, I accept that the Transgenic Core can not guarantee that targeted clones will be produced. If no targeted clones result, we will reconsider the design of the experiment. This is because of the inherent variability in homologous recombination in embryonic stem cells. I understand that it is expected that the expertise and effort invested in this project by Elizabeth Hughes and Thomas Saunders will be recognized appropriately. I agree to pay the University of Michigan approved recharge for this service. The Transgenic Core will undertake to produce gene targeted embryonic stem cells. The Core will provide/perform the following information/procedures: Prepare an experimental time line for planning purposes. Electroporate the targeting vector into ___ ES cells. Will pick 480 electroporated ES cell clones (five 96-well plates). Each plate of the five 96-well plates will be split into three. Two plates will be cryopreserved in independent -80°C freezers. One plate will be grown and split into two replicates for DNA preparation. Prepare DNA from replicate 96-well plates (ten plates total, two replicates of each of the five plates of clones). Deliver the DNA plates to the investigator for screening. Authorized Signature Date Core Representative Signature Date The University of Michigan Transgenic Animal Model Core Sally A. Camper, Ph.D. Linda C. Samuelson, Ph.D. Thomas L. Saunders, Ph.D. 2560 MSRB II, Box 0674 1150 W. Medical Center Drive Ann Arbor, MI 48109-0674 Voice: (734) 647-2910 FAX: (734) 936-2622 http://www.med.umich.edu/tamc/ Production of Gene Targeted Embryonic Stem Cell Clones Statement of Understanding of the Nature of the Gene Targeting Service For production of gene targeted embryonic stem cells in collaboration with the Transgenic Core I am responsible for the following tasks: Restriction map the endogenous gene. Establish a robust, reliable, reproducible screen that discriminates between wild type and targeted alleles. Provide experimental data demonstrating that the Southern blot probes will confirm the genomic structure of targeted ES cell clones. Clone the gene targeting vector. Purify gene targeting vector DNA for electroporation according to the protocol supplied by the Transgenic Core. Analyze genomic DNA from as many as 480 ES cell clones for homologous recombination with the targeting vector. Timely analysis is important because of the limited life span of ES cells cryopreserved at 80°C. Review the screening data with the Transgenic Core prior to expansion of cryopreserved clones. Although 480 clones should contain multiple targeted clones, I accept that the Transgenic Core can not guarantee that targeted clones will be produced. If no targeted clones result, we will reconsider the design of the experiment. This is because of the inherent variability in homologous recombination in embryonic stem cells. I understand that it is expected that the expertise and effort invested in this project by Elizabeth Hughes and Thomas Saunders will be recognized appropriately. I agree to pay the $4,000 recharge for this service. The Transgenic Core will undertake to produce gene targeted embryonic stem cells. The Core will provide/perform the following information/procedures: Prepare an experimental time line for planning purposes. Electroporate the targeting vector into ___ ES cells. Will pick 480 electroporated ES cell clones (five 96-well plates). Each plate of the five 96-well plates will be split into three. Two plates will be cryopreserved in independent -80°C freezers. One plate will be grown and split into two replicates for DNA preparation. Prepare DNA from replicate 96-well plates (ten plates total, two replicates of each of the five plates of clones). Deliver the DNA plates to the investigator for screening. Authorized Signature Date Core Representative Signature Date The University of Michigan Transgenic Animal Model Core Sally A. Camper, Ph.D. Linda C. Samuelson, Ph.D. Thomas L. Saunders, Ph.D. 2560 MSRB II, Box 0674 1150 W. Medical Center Drive Ann Arbor, MI 48109-0674 Voice: (734) 647-2910 FAX: (734) 936-2622 http://www.med.umich.edu/tamc/ Billing Information Project Name Date Lab Contact Phone Number Preferred form of contact: email address FAX Number P.I. Name Authorized Signer: Authorized Signature: Phone Number Shortcode Number Billing Department Grant Title: Funding Agency: Grant Number: The University of Michigan Transgenic Animal Model Core Thomas L. Saunders, Ph.D. Elizbeth D. Hughes, M.S. 2560 MSRB II, Box 0674 1150 W. Medical Center Drive Ann Arbor, MI 48109-0674 TEL: (734) 647-2910 FAX: (734) 936-2622 http://www.med.umich.edu/tamc/ Purification of Gene Targeting Vector DNA for Electroporation 1. Purify plasmid from bacteria. We recommend the either the Sigma-Aldrich GenElute HP Endotoxin Free Plasmid Purification kit or the Qiagen EndoFree Plasmid Maxi kit for the purification of the targeting vector plasmid from bacteria. Please follow the directions in the kit. Electroporation of Qiagen purified DNA has been used successfully by a number of labs. Alternatively, plasmid DNA can be purified by CsCl banding. 2. Provide 200 µg of linearized plasmid DNA to the Transgenic Core. Use a restriction enzyme that cuts once in the plasmid vector. Run a little DNA on a minigel to verify that digestion is complete. Extract the DNA with one volume phenolchloroform, then with one volume chloroform and precipitate by adding 0.1 volume 5M NaCl and 2 volumes cold ethanol. Resuspend the DNA in sterile TE (10 mM Tris-HCl, pH 8.0, 1.0 mM EDTA) at 1 mg/ml and deliver it to the Transgenic Core for electroporation. Prior to electroporation, we will verify the concentration and run it on a minigel to check the size. --------------------------------------------------------------------Please provide the following information when you submit the targeting vector. Name of Targeting Vector Size of linearized Vector DNA Resuspended in: DNA Concentration: Date: Contact Person: phone/email: