Supplementary Data
advertisement

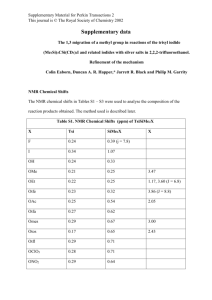
Supplementary Data Figure 1: Distribution of criteria for automated removal of 97% of detected peak groups by the default IDEOM filters. Of 27,727 peak groups (m/z and retention time pairs), 82 were assigned with confident metabolite identifications based on authentic standards, 506 were putatively identified based on m/z and retention time and 373 unidentified peak groups remained after noise filtering and identification procedures. Full details of IDEOM filtering procedures are recorded in the IDEOM documentation (http://mzmatch.sourceforge.net/ideom/Ideom_Documentation.pdf). Noise filter parameters applied to this study are listed in supplementary table 1. Filtering annotations and confidence scores for all individual peak groups are recorded in the ‘alldata’ worksheet of the IDEOM Excel file for this study: [insert weblink here]. NOTE: the total number of peaks is slightly higher than usually detected in untargeted metabolomics studies because ‘greedy’ peak-picking parameters were used in an attempt to improve detection of small isotopomer peaks. Nevertheless, the noise filters appeared to effectively remove the excess noise from the dataset for metabolite identification. Unidentified: Probable MS artefacts 7% Contaminants or Background 22% Other Filtered (inc. Rt & ppm) 2% Small or Variable peaks 11% Metabolites 3% MS artefacts 27% LC noise or Peakpicking artefacts 28% Condifident identifications 0.3% Putative identifications 2% Unidentified possible metabolites 1% Table 1: Parameters applied for data processing with XCMS, mzMatch and IDEOM. XCMS Method: ppm: peak width (min): peak width (max): S/N threshold: Prefilter (# points): Prefilter (intensity): Mzdiff: normal mzXML 2 5 seconds 100 seconds 2 2 500 0.001 mzMatch Mzmatch grouping RT window: Mzmatch grouping m/z ppm: 0.5 min 5 ppm Relative Std Dev (RSD) filter: Noise filter (codadw): Intensity filter (LOQ): Minimum detections # RT window for related peaks: 1 0.5 500 2 0.1 min IDEOM Relative Std Dev (RSD) filter: Noise filter (codadw): Intensity filter (LOQ): Minimum detections # RT window for related peaks: RT for id of authentic standards: RT for id for calculated RT: PPM for mass identification: PPM after re-calibration Ignore related peaks before RT: RT window for complex adducts: RT window for Duplicatepeaks: RT window for Shoulderpeaks: Intensity ratio for Shoulderpeaks: 0.25 0.75 5000 3 0.2 10 45 5 3 0 0.5 1 2 5 STRICT min % % ppm ppm min min min min to 1 (for ratio Minimum detectable intensity: 1000 calc) 0.05 Unpaired Statistical P-value: Preferred DB: TrypDB tbr File 1: (Suppfile1.xlsx) Excel spreadsheet summary of identified and putatively identified metabolites in procyclic-form Trypanosoma brucei. 13C-labelled metabolites are annotated in column J. Detailed peak intensities for each metabolite and isotopomer are listed in the ‘RAWisotopomerPEAKS’ sheet. All data from this study, and full functionality for data analysis and visualisation, are available in the IDEOM file (insert weblink here) File 2: (isotopes_091013080512_neg.pdf) PDF file containing all mass traces and graphs for isotopomers of putatively identified metabolites from negative mode MS ionisation. File 3: (isotopes_142459070512_pos.pdf) PDF file containing all mass traces and graphs for isotopomers of putatively identified metabolites from positive mode MS ionisation.