here - Helmholtz Zentrum München
advertisement

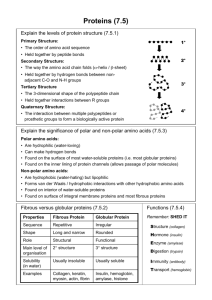
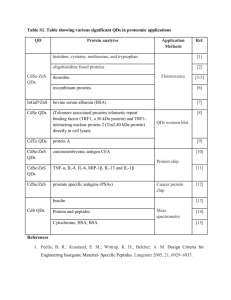
NMR tools for studying protein complexes Steffen Glaser1, Bernd Reif1,2, Michael Sattler1,2 1 Institute of Structural Biology, Helmholtz Zentrum München, 85764 Neuherberg, Germany 2 Center for Integrated Protein Science Munich (CiPSM) at Biomolecular NMR, Department Chemie, Technische Universität München, 85747 Garching, Germany We are studying the structures, molecular interactions and posttranslational modifications of proteins and protein complexes. Structural analysis of these posttranslational modifications also required the use of optimized isotope labelling and NMR methods, which will be presented. For structural analysis of we have developed a versatile and efficient protocol for determining the quaternary structure of multi-domain proteins and protein complexes in solution by combining experimental data derived from solution state NMR as well as Small Angle X-ray and/or Neutron Scattering (SAXS/SANS) experiments [2-4]. Information about the relative orientation of domains or subunits is obtained from NMR residual dipolar couplings (RDCs). Long-range (up to 20Å) distance restraints are obtained from paramagnetic relaxation enhancements (PRE) using spinlabeled proteins and/or RNA. References: [1] C.D. Mackereth, M. Sattler, Curr. Op. Struct. Biol 2012, 22:287-96 [2] F. Gabel, et al , J Biomol NMR 2008, 41, 199. [3] B. Simon, et al Angew Chem Int Ed Engl 2010, 49, 1967. [4] Madl T, Gabel F, and Sattler M J Struct Biol 2011, 173, 472-82.