Supplementary Information (doc 56K)
advertisement

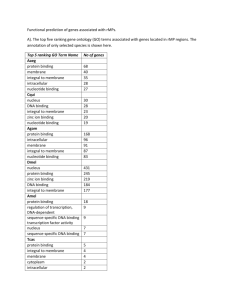
The Mutational Pattern of Primary Lymphoma of the Central Nervous System Determined by Whole Exome Sequencing Running Title: Mutational Pattern of PCNSL Inga Vater*1, Manuel Montesinos-Rongen*2, Matthias Schlesner*3, Andrea Haake1, Frauke Purschke2, Rosanne Sprute2, Nina Mettenmeyer2, Ilias Nazzal2, Inga Nagel1, Jana Gutwein1, Julia Richter1, Ivo Buchhalter3, Robert B. Russell4,5, Otmar D. Wiestler6, Roland Eils3,5,7, Martina Deckert**2, Reiner Siebert**1 1Institute of Human Genetics, Christian-Albrechts-University Kiel & University Hospital Schleswig-Holstein, Campus Kiel, Kiel Germany 2Institute 3Division of Neuropathology, University of Cologne, Cologne, Germany of Theoretical Bioinformatics (B080), German Cancer Research Center (DKFZ), Heidelberg, Germany 4Cell Networks, University of Heidelberg, Heidelberg, Germany 5BioQuant, 6German Heidelberg University, Heidelberg, Germany Cancer Research Center (DKFZ), Heidelberg, Germany 7Department for Bioinformatics and Functional Genomics, Institute for Pharmacy and Molecular Biotechnology (IPMB), Heidelberg University, Heidelberg, Germany *These authors contributed equally to this work ** These authors contributed equally to this work Tables Supplementary Table 1. Primers Supplementary Table 2. Sequence variants (after filtering) Supplementary Table 3. Genes recurrently affected by protein changing variants Figure Legends Supplementary Figure 1. Verification of the Whole Exome Sequencing Method Supplementary Figure 2. Expression of the recurrently mutated genes mining on gene expression profiling of PCNSL Supervised analysis of PCNSL vs. DLBCL for all mutated genes, which affected at least two PCNSL, significantly differentially expressed tags were calculated based on Student's t-test. Only tags that were at least 2-fold significantly differentially expressed are shown. Samples of normal brain tissue were included for comparison. 2 Supplementary References 1. Malinauskas T, Aricescu AR, Lu W, Siebold C, Jones EY. Modular mechanism of Wnt signaling inhibition by Wnt inhibitory factor 1. Nature structural & molecular biology 2011 Aug; 18(8): 886-893. 2. Minet AD, Rubin BP, Tucker RP, Baumgartner S, Chiquet-Ehrismann R. Teneurin-1, a vertebrate homologue of the Drosophila pair-rule gene ten-m, is a neuronal protein with a novel type of heparin-binding domain. Journal of cell science 1999 Jun; 112 ( Pt 12): 2019-2032. 3. Dinkel H, Van Roey K, Michael S, Davey NE, Weatheritt RJ, Born D, et al. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res 2014 Jan; 42(Database issue): D259-266. 4. Schiefner A, Gebauer M, Skerra A. Extra-domain B in oncofetal fibronectin structurally promotes fibrillar head-to-tail dimerization of extracellular matrix protein. J Biol Chem 2012 May 18; 287(21): 17578-17588. 5. Almo SC, Bonanno JB, Sauder JM, Emtage S, Dilorenzo TP, Malashkevich V, et al. Structural genomics of protein phosphatases. Journal of structural and functional genomics 2007 Sep; 8(2-3): 121-140. 6. Appleton BA, Wu P, Maloney J, Yin J, Liang WC, Stawicki S, et al. Structural studies of neuropilin/antibody complexes provide insights into semaphorin and VEGF binding. The EMBO journal 2007 Nov 28; 26(23): 4902-4912. 7. Forneris F, Ricklin D, Wu J, Tzekou A, Wallace RS, Lambris JD, et al. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science 2010 Dec 24; 330(6012): 18161820. 3 4