emi412321-sup-0003-si
advertisement

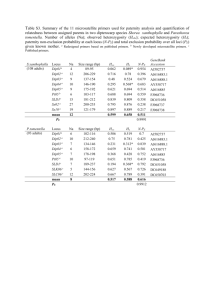
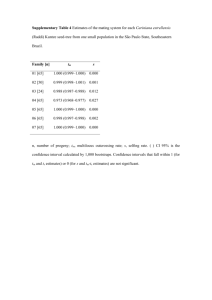
Appendix S1. Experimental Procedures Fungal isolates and DNA extraction The fungal isolates were retrieved from primary cultures initiated from a single selected colony, having previously been identified on the basis of macroscopic and microscopic morphological characteristics and standard mycological procedures. Subsequently, identification was confirmed by sequencing analyses of β-tubulin and rodlet A genes and microsatellite typing (Serrano et al., 2011; Araujo et al., 2012). Before DNA extraction, the fungi were grown for five days on Sabouraud dextrose agar slants (Difco) at 30 ºC. DNA (50-250 ng) was extracted from conidia using the sodium hydroxide method (http://www.aspergillus.org.uk/indexhome.htm?secure/laboratory_ protocols) and it was suspended in 50 µL of sterile water and stored at -20 ºC. DNA amplification and sequencing Previously described primers for MLST amplification (Caramalho et al., 2013) and microsatellite-multiplex (Araujo et al., 2009) were employed and the remaining primers designed using Primer3 v.0.4.0 software (http://bioinfo.ut.ee/primer3-0.4.0/). Primerdimer and hairpin structures were avoided by using AutoDimer v.1.0 (http://www.cstl.nist.gov/biotech/strbase/AutoDimerHomepage/AutoDimerProgramHo mepage.htm). The list of primers is available on supplementary Table S1. The primers were initially tested in singleplex reactions for confirmation of its specificity. Then, multiplex PCR was performed in a total reaction volume of 5 µL, as previously described (Caramalho et al., 2013). The amplification products were confirmed on polyacrylamide gel and visualised through standard silver-staining (Qu et al., 2005). Multiplex PCR products were purified with ExoSAP-IT (USB Corporation) 1 and sequencing reactions performed as previously described (Araujo et al., 2009). Afterwards the reactions were analysed employing Sequencing 5.2 analysis software (Applied Biosystems). Mini-sequencing SNaPshot assays Single base extension (SBE) primers were designed using the previously referred AutoDimer v.1.0 software (see supplementary information Figure S1). Two minisequencing reactions were set up using 1.5 µL of purified multiplex PCR product, 1.0 µL of SBE primers mix, 1.0 µL of SNaPshot® Multiplex Reaction Ready Mix (Applied Biosystems) and 1.5 µL of ultrapure water. The cycling conditions were 25 cycles at 96 ºC for 10 s, 50 ºC for 5 s, and 60 ºC for 30 s (Eusebio et al., 2013). Unincorporated ddNTPs were eliminated with SAP (USB) and purified products were mixed with 9.0 µL of HiDiTM formamide (Applied Biosystems) and GeneScan-120 LIZ size standard. Capillary electrophoresis was performed in ABI 3130xl Genetic Analyser (Applied Biosystems) and the results of mini-sequencing analysed with GeneMapper® v.4.0 (Applied Biosystems). Linkage disequilibrium analyses The index of association (Ia) and rBarD values were calculated using Multilocus 1.3b available at http://www.agapow.net/software/multilocus/ (Agapow and Burt, 2001). Ia gives information of linkage disequilibrium allowing the comparison of individuals that share the same allele at one locus and evaluating the possibility of sharing the same allele at other loci. Ia is equal to zero if there is no linkage disequilibrium and this value increases when strong linkage disequilibrium is found. The null hypothesis of complete 2 panmixia (Ia = 0) was tested with Multilocus 1.3b, by comparing the observed data set after 1,000 randomizations. References Agapow, P.M., and Burt, A. (2001) Indices of multilocus linkage disequilibrium. Mol Ecol Notes 1: 101–102. Araujo, R., Pina-Vaz, C., Gonçalves Rodrigues, A., Amorim, A., and Gusmão, L. (2009) Simple and highly discriminatory microsatellite-based multiplex PCR for Aspergillus fumigatus strain typing. Clin Microbiol Infect 15: 260-266. Araujo, R., Amorim, A., and Gusmão, L. (2012) Diversity and specificity of microsatellites within Aspergillus section Fumigati. BMC Microbiol 12: 154. Caramalho, R., Gusmão, L., Lackner, M., Amorim, A., and Araujo, R. (2013) SNaPAfu: A novel single nucleotide polymorphism multiplex assay for Aspergillus fumigatus direct detection, identification and genotyping in clinical specimens. PLoS One 8: e75968. Eusebio, N., Pinheiro, T., Amorim, A., Gamboa, F., Saraiva, L, Gusmão L., et al. (2013) SNaPaer: a practical single nucleotide polymorphism multiplex assay for genotyping of Pseudomonas aeruginosa. PLoS One 8: e66083. Qu, L., Li, X., Wu, G., and Yang, N. (2005) Efficient and sensitive method of DNA silver staining in polyacrylamide gels. Electrophoresis 26: 99-101. Serrano, R., Gusmao, L., Amorim, A., and Araujo, R. (2011) Rapid identification of Aspergillus fumigatus within the section Fumigati. BMC Microbiol 11: 82. 3