tpj12408-sup-0012-MethodS2
advertisement

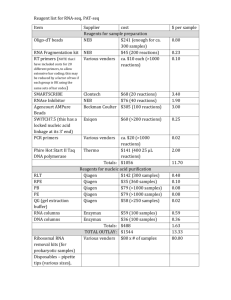
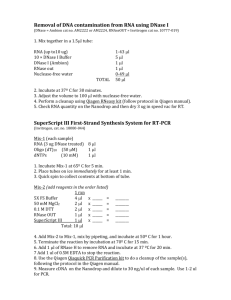
Method S2. RT-PCR Total RNA was isolated using a combined TRIzol and silica spin column protocol. Approximately 100 mg frozen plant tissue was mixed with 1 ml of TRIzol (Invitrogen) and incubated for 5 min at room temperature. Subsequently, samples were mixed vigorously with 200 µl of chloroform and centrifuged at 12,000 g for 10 min at 4 C. The aqueous phase was collected and used for RNA isolation with the Qiagen RNeasy column according to the manufacturer’s instructions. On-column DNase digestion was carried out using RNase-Free DNase set (Qiagen). 1 µg of total RNA was used for cDNA synthesis with SuperScript III Reverse Transcriptase (Invitrogen), following the manufacturer’s recommendations. For semiquantitative RT-PCR, a 1 μl aliquot of the 20 μl cDNA reaction mixture was used as a template. Transcript levels of CYB (At1g57620) were analyzed after 35 cycles, using primers Forward (F): 5’ CTGTACCTCACACCGGATCA 3’ and Reverse (R): 5’ TGGTGTAACACGGTGCCATA 3’. Transcript levels of GLL23 were analyzed after 30 cycles, using primers F: 5’ GAACGAATTGGCTAGAACAG 3’ and R: 5’ GCAACCATAAGCATCATGTG 3. Actin 8 was used as a reference gene (primers F: 5’ GAGACATCGTTTCCATGACG 3’; R: 5’ TTTCAAACCTGCTCCTCCTT 3’). Real-time qRT-PCR was performed in a Bio-Rad iQ5 thermal cycler. For the PCR amplification with GLL23- and ACT8-specific primers (sequences as above), iQ SYBR Green Supermix (BioRad) was used according to the manufacturer’s instructions in a final volume of 20 μl. The fold change in gene expression was calculated using the CP equation of Pfaffl (2001). References to Method S2 Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. 1