Experiment 2: UV-Vis and Fluorescence
advertisement

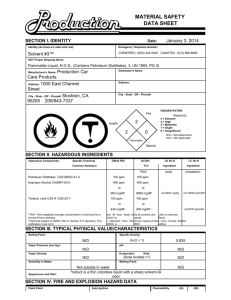
UV-Vis and Fluorescence Introduction: A UV-Vis spectrophotometer is an instrument used to analyze compounds in ultraviolet and visible regions of the electromagnetic spectrum. It can also be used to determine wavelength and the maximum absorbance of compounds. It does this by looking at electronic transitions. The spectrophotometer we will be using is the Thermo Evolution 600 UV-Vis. Fluorescence is used to determine the quantitative measurements of an analyte by using two wavelengths along the spectrum. It can find trace concentrations as low as 1 ppt. A fluorescence instrument sends light into a sample and excites the electrons. Then, it reads the radiation released when the electrons return to their ground states. The instruments we will be using are the Shimadzu Spectrofluorophotometer and the PE Luminescence Spectrophotometer. Purpose: The purpose of this lab is to determine the absorbance levels of Salicylic Acid (SA) in aged aspirin samples. This will be done by running samples of SA in hexanes through the UVVis instrument. We will also be determining the amount of quinine in tonic water through the fluorescence instruments. Procedure: UV-Vis 1. Prepare standard solutions of 40, 25, 10, 7, and 2 ppm from 1000 ppm stock solution of SA in hexane. 2. Run 3 samples of each standard on instrument. 3. Create calibration curves from data. 4. Dissolve and filter an aspirin tablet through hexane. Run new solution on instrument. Instrument Use 1. Power on instrument (switch is on left side). 2. Turn on monitor and double-click ‘VisionPro’ a. If not logged in, username is student and there is no password 3. Instrument will run through a start-up check for about 10-15 minutes. 4. In the toolbar frame, select the Scan icon. 5. In dialog box, enter wavelength from 190-400 nm. a. Choose Absorbance and Tungsten Lamp. 6. Click on Baseline Correction. a. Make sure 100% T is selected. 7. Click on Baseline/Zero icon. 8. Wash a quartz cuvette with hexane three times, then fill ¾ of the way with sample and dry with chem wipes. 9. Place cuvette in the first slot with the frosted sides facing you and the window. 10. Click the Run icon. 11. To obtain peak and absorbance, click on the graph window, then go to the graph menu at the top and choose Peak Labels and select Wavelength and Value. a. In the menu graph again, select Track at the top of the list and click on Graph. b. Using arrow keys, find the peak around 310 nm and record the absorbance. Fluorescence 1. Prepare standards of 2,3,4, and 5 ppm solutions of quinine in 0.1M H2SO4 using 1000 ppm stock solution. 2. Run three samples of each concentration through the fluorescence instruments and record the wavelength and intensity of the largest peak. 3. Make a solution of tonic water by diluting 5 mL tonic water with 95 mL H2SO4. 4. Run three scans of solution through both instruments and record the wavelength and intensity of largest peak. Instrument Use: Shimadzu 1. Turn on instrument and computer. 2. Open PX-150x program on computer. 3. Check lower right corner to see if instrument is on. a. If not, go to Configure Menu and go down to PC Configuration to make sure First Portal is selected. b. Then, under Configure Menu, go to the Instrument and make sure On is selected. 4. Go to Acquire Mode and select Spectrum. 5. Make sure standards are prepared. 6. Fill cuvette with solution and run under these settings: a. Spectrum type: excitation b. EM wavelength: 400 nm c. Ex wavelength range: 220 (start), 900 (end) d. Sensitivity: Low e. Scan speed: Fast f. Recording range: -10.00 (low), 500.00 (high) g. EX/EM slit width: 10/10 h. Response time: 0.02 i. Repeat Scan/Auto File: No setting needed 7. Click Search λ and then click Search. a. Ensure the default range for EX (excitation) and EM (emission) are 230-450 nm and 240-650 nm. 8. Reset parameters to this: a. Spectrum type: emission b. EX wavelength: (ideal wavelength found) c. EM wavelength range: (include optimal EM wavelength) d. Rest of the parameters are the same as before 9. Open left hatch and pull out platform to insert cuvette. Hit Start. 10. Run scan of each concentration and record wavelength and intensity of largest peak. a. In order to see the peaks, click Manipulate then Peak Pick. 11. Run scan of tonic water solution and record wavelength and intensity of largest peak. Instrument Use: Perkin Elmer 1. Turn on instrument. 2. Open WINLab and file Default.MTH 3. Fill cuvette with blank (H2SO4) and run a scan with the same parameters as before except change the slit from 8 to 3 for both. 4. Run samples three times. 5. Record wavelength and intensity of largest peak. a. Click button of a peak with a vertical line through it and drag line to the middle of the largest peak. b. Data found towards the bottom of the screen. 6. Repeat for tonic samples. Data: UV-Vis PPM 2 7 10 25 40 Wavelength 311 309 309 313 313 Absorbance 0.157 0.284 0.386 1.038 1.366 UV/VIS y = 0.0336x + 0.082 R² = 0.9827 Absorbance 1.5 1 0.5 0 0 5 10 15 20 25 Concentration (ppm) 30 35 40 45 Fluorescence Shimadzu Excitation: 344nm Emission: 451nm 2 ppm Trial Wavelength (nm) Intensity 1 446 446.298 2 447 474.997 3 448 485.882 Avg. 447 469.059 Trial Wavelength (nm) Intensity 1 448 699.102 2 447 683.645 3 449 710.639 Avg. 448 697.795 Trial Wavelength (nm) Intensity 1 449 940.873 2 447 950.862 3 446 951.760 Avg. 447.3 947.832 3 ppm 4 ppm 5 ppm – For all three trials, the data was beyond the range of the instrument. Therefore, no data was collected for this sample. Weis Trial Wavelength (nm) Intensity 1 449 873.556 2 447 882.043 3 448 877.798 Avg. 448 877.799 Trial Wavelength (nm) Intensity 1 450 435.836 2 450 439.123 3 448 437.492 Avg. 449.3 437.484 Giant Intensity Shimazdu Spectrofluorophotometer 1000 900 800 700 600 500 400 300 200 100 0 y = 239.39x - 13.264 R² = 0.9993 0 0.5 1 1.5 2 2.5 Concentration (ppm) 3 3.5 4 4.5 Perkin Elmer ppm 0.00 2.00 3.00 4.00 5.00 Weis Giant Wavelength (nm) 339.43 443.00 443.94 444.45 beyond range 443.43 445.21 Intensity 61.98 513.95 741.20 970.30 beyond range 901.94 465.87 Perkin Elmer 1200.00 Intensity 1000.00 800.00 600.00 y = 226.97x + 61.176 R² = 1 400.00 200.00 0.00 0.00 0.50 1.00 1.50 2.00 2.50 3.00 3.50 Concentration (ppm) Calculations: UV-Vis 1000 ppm = .1 g/ 100 mL Salicylic Acid Standards 40 ppm (40 ppm)(50 mL) = (1000 ppm)(V) V = 2 mL 25 ppm (25 ppm)(50 mL) = (1000 ppm)(V) V = 1.25 mL 10 ppm (10 ppm)(50 mL) = (1000 ppm)(V) V= 0.5 mL 4.00 4.50 7 ppm (7 ppm)(50 mL) = (1000 ppm)(V) V= 0.35 mL 2 ppm (2 ppm)(50 mL) = (1000 ppm)(V) V = 0.1 mL Fluorescence Dilution to 100 ppm Stock Solution (1000 pppm Stock)(V) = (100 ppm)(100 mL) V= 10 mL M H2SO4 (0.1 M)(500 mL) = (18 M)(V) V = 2.78 mL Standards 2 ppm (2 ppm)(100 mL) = (100 ppm)(V) V= 2 mL 3 ppm (3 ppm)(100 mL) = (100 ppm)(V) V= 3 mL 4 ppm (4 ppm)(100 mL) = (100 ppm)(V) V= 4 mL 5 ppm (5 ppm)(100 mL) = (100 ppm)(V) V= 5 mL Conclusion: This week we successfully used the UV-Vis and Fluorescence instruments. On the first day, we ran standard concentrations on the Shimadzu and Perkin Elmer Fluorescence instruments. After plotting the concentration vs. the intensity on a calibration curve for the Shimadzu data, our R squared value 0.9993. This proves we had very good results. We also had good results for the Perkin Elmer since our R squared value for the data was 1. There was no unknown given to be tested on the fluorescence instruments. On the second day, we ran standards on the UV-Vis instrument. For this instrument, we created our own standards and stock solution. After plotting the concentration vs. the absorbance on a calibration curve, our R squared value for the data was 0.9827. Although this is not as good as our other R squared values for the fluorescence instruments, it still proves our experiment was a success. There were several problems with this lab. First, there were no samples to run for the Fluorescence instrument. Also, the instructions for the Perkin Elmer and the Shimadzu instruments were not very clear and were missing some important steps. For the UV/Vis experiment, the aspirin tablets did not dissolve after stirring and heating for two hours so no samples were run. Overall, our experiment was a success and we were able to collect good data from all three instruments.