Supplementary Figure Legends (doc 38K)
advertisement

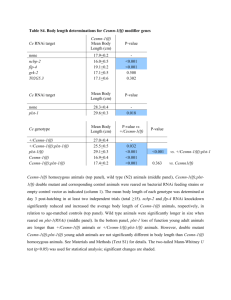
Supplementary Figure Legends Figure S1. Validation of the ChIP-on-chip results by ChIP-qPCR. (a) After post-confluence, cells were induced to differentiation. ChIP-qPCR was performed at 20 h after induction with C/EBP antibody (sc-150) and control IgG. C/EBPβ enrichment was assayed at 30 novel C/EBPβ binding sites as listed in Supplementary Table S1. The 30 sites were selected to represent a range of fold enrichment values based on signal intensity in the ChIP-on-chip result. One region of the insulin gene serves as a negative control. Primer sequences for the 30 novel C/EBPβ binding sites in the ChIP-qPCR assays are available upon request. 29 of these sites showed truely positive, except CEBPB_374, as indicated by #. (b) C/EBPβ enrichment at 15 of the 30 C/EBPβ binding sites was tested by using two different antibodies against C/EBPβ (sc-150 and sc-7962). Figure S2. EMSA for C/EBPβ with Kdm4b promoter. EMSA was performed with a 32 P-labeled probe containing the C/EBP binding site (-577 to -564 bp) of Kdm4b promoter and nuclear extracts from 3T3-L1 cells at 20 h after induction. Supershift experiment was performed with control IgG or C/EBPβ antibody. For competition analysis, unlabeled competitor probe of the C/EBP element in the C/EBP α gene promoter (Consensus) or unlabeled Kdm4b self probe mutated in the C/EBP element (Mut) were used. The positions of the C/EBPβ containing DNA-protein complex and the supershift band (SS) are indicated. Figure S3. Kdm4b RNAi has no effect on the expression of the other Kdm4 family genes. 3T3-L1 cells were transfected with control RNAi or Kdm4b RNAi. The expression of the indicated genes was detected by RT-qPCR. Data normalized to control RNAi group of each gene. *p < 0.05. Figure S4. Promoters of the four cell cycle genes identified in the ChIP-on-chip data are indicated. The C/EBPβ consensus sequences and the mutated sequences are shown below. Figure S5. RNAi-mediated knockdown of the four cell cycle genes inhibits MCE and adipogenesis. (a) 3T3-L1 preadipocytes were transfected with control RNAi or RNAi directed against the four cell cycle genes and then induced to differentiation. Cell lysates were obtained at 20 h and the expression level of the indicated genes was determined by Western blotting. (b) 3T3-L1 preadipocytes were treated with the indicated RNAi. At 18 h after induction, cells were labeled with EdU for 2 hrs and then subjected to flow cytometry analysis. The percentages of cells in S phase were determined. (c) At 18 h after induction, cells were labeled with EdU for 2 hrs and then stained with Hoechst. The fluorescence of EdU (red) and Hoechst (blue) was detected with a fluorescence microscope. (d) Cell numbers were determined at times indicated after induction. Data normalized to 0 h time point. (e) At 8 d after induction, cells were stained with Oil red O. (f) At 8 d after induction, the mRNA levels of adipocyte marker genes were determined by RT-qPCR. Figure S6. Over-expression of the four cell cycle genes together significantly rescues the inhibition of MCE mediated by C/EBPβ RNAi knockdown. 3T3-L1 preadipocytes were infected with empty vector or co-infected with retroviruses expressing Cdc45l, Mcm3, Gins1 and Cdc25c. Then cells were transfected with control RNAi or C/EBPβ RNAi. After post-confluence, cells were induced to differentiation. (a) Cells were harvested at 20 h to detect the expression of the indicated genes by Western blotting. (b) EdU incorporation assay was conducted. (c) Cell number counting was performed. (d) At 8 d after induction, cells were stained with Oil red O. (e) At 8 d after induction, the mRNA levels of adipocyte marker genes were determined by RT-qPCR. *p < 0.05. Figure S7. Proposed model: the mechanistic explanation for the role of C/EBPβ during MCE and terminal adipocyte differentiation in 3T3-L1 cells. Figure S8. Overlap of binding regions between C/EBPβ and Kdm4b at 20 h of 3T3-L1 adipocyte differentiation. Shown in parentheses is the number of enriched regions for each antibody. Figure S9. Kdm4b regulates the expression of histone H4 through demethylation of H3K9me3. (a) ChIP analysis indicated that Kdm4b associated with the promoter of histone H4 at 20 h post-induction. (b) Decrease of H3K9me3 modification at the promoter of histone H4 was compromised by Kdm4b RNAi. 3T3-L1 preadipocytes were treated with control RNAi or Kdm4b RNAi and then induced to differentiation. Cells were collected at times indicated and subjected to ChIP-qPCR analysis of H3K9me3 modification at the promoter of histone H4. Data was normalized to 0 h time point of control RNAi-treated cells. (c) Kdm4b was required for histone H4 expression during MCE. 3T3-L1 preadipocytes were treated as in (b). The relative mRNA level of histone H4 at times indicated was analyzed by RT-qPCR.