Galaxy-History_1-Unstable and Stable Region Derivation based on
advertisement

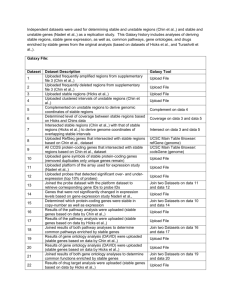
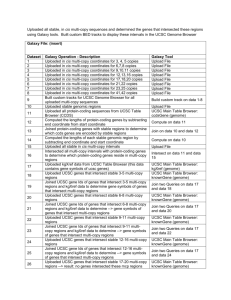
Merged genome coordinates pertaining to probes that detected the same copy number abnormality to create larger contiguous intervals of unstable regions. Used Galaxy tools to determine stable regions and coding genes associated with each stable or unstable region. Galaxy file: (insert) Dataset 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 Dataset Description Uploaded CCDS gene track from UCSC table browser (coding consensus genes) Uploaded frequently deleted probes (data from Hicks et al., supplementary file) Uploaded frequently amplified probes (data from Hicks et al., supplementary file) Compared the amplified/deleted probe sets --> probes that were similar to both sets Compared the amplified/deleted probe sets --> probes that are unique to amplified set Compared the amplified/deleted probe sets --> probes that are unique to deleted set Uploaded file that numerically ordered probe Ids and annotated each by: del, amp, or both Uploaded file containing clustered coordinates of the probes that belonged to detecting the same copy number abnormality - i.e. Coordinates of unstable regions Complemented the unstable regions to derive genomic coordinates of stable regions Intersected CCDS genes and unstable regions to get --> coding genes in unstable regions Intersected CCDS genes and stable regions to get --> coding genes in stable regions Determined level of coverage for each coding gene in stable region (percentage of coverage is in column 8 of table) Built custom track to display unstable regions in UCSC Genome Browser Built custom track to display stable regions in UCSC Genome Browser Uploaded genomic coordinates of clustered intervals based on probes that detected amplifications Uploaded genomic coordinates of clustered intervals based on probes that detected both amplifications and deletions Uploaded genomic coordinates of clustered intervals based on probes that detected deletions Intersected coding genes and amplified regions Intersected coding genes and deleted regions Intersected coding genes and both deleted/amplified regions Uploaded file containing coding genes that are 100% covered by stable genomic regions Uploaded 812 stable regions Built custom track for UCSC Genome Browser based on stable region coordinates Galaxy Tool UCSC Main Table Browser: knownGene (genome) Upload File Upload File Compare two Queries on data 2 and data 3 Compare two Queries on data 2 and data 3 Compare two Queries on data 2 and data 3 Upload File Upload File Complement on data 8 Intersect on data 1 and data 8 Intersect on data 1 and data 9 Coverage on data 9 and data 11 Build custom track on data 8 Upload File Upload File Upload File Upload File Intersect on data 1 and data 15 Intersect on data 1 and data 17 Intersect on data 1 and data 16 Upload File Upload File Build custom track on data 22