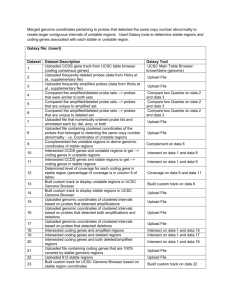
Galaxy-History_6-Unstable and Stable Region Derivation based on
advertisement

Independent datasets were used for determining stable and unstable regions (Chin et al.,) and stable and unstable genes (Naderi et al.,) as a replication study. This Galaxy history includes analyses of deriving stable regions, stable gene expression, as well as, common pathways, gene ontologies, and drugs enriched by stable genes from the original analysis (based on datasets of Hicks et al., and Turashvili et al.,). Galaxy File: Dataset 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 Dataset Description Uploaded frequently amplified regions from supplementary file 3 (Chin et al.,) Uploaded frequently deleted regions from supplementary file 3 (Chin et al.,) Uploaded stable regions (Hicks et al.,) Uploaded clustered intervals of unstable regions (Chin et al.,) Complemented on unstable regions to derive genomic coordinates of stable regions Determined level of coverage between stable regions based on Hicks and Chins data Intersected stable regions (Chin et al.,) with that of stable regions (Hicks et al.,) to derive genome coordinates of overlapping stable intervals Uploaded RefSeq genes that intersected with stable regions based on Chin et al., dataset All CCDS protein-coding genes that intersected with stable regions based on Chin et al., dataset Uploaded gene symbols of stable protein-coding genes (removed duplicates only unique genes remain) Uploaded platform of the array used for expression study (Naderi et al.,) Uploaded probes that detected significant over- and underexpression (top 10% of probes) Joined the probe dataset with the platform dataset to retrieve corresponding gene IDs to probe IDs Genes that were not significantly changed in expression levels based on gene expression study Naderi et al., Determined which protein-coding genes were stable in copy-number as well as expression Results of the pathway analysis were uploaded (stable genes based on data by Chin et al.,) Results of the pathway analysis were uploaded (stable genes based on data by Hicks et al.,) Joined results of both pathway analyses to determine common pathways enriched by stable genes Results of gene ontology analysis (DAVID) were uploaded (stable genes based on data by Chin et al.,) Results of gene ontology analysis (DAVID) were uploaded (stable genes based on data by Hicks et al.,) Joined results of both gene ontology analyses to determine common functions enriched by stable genes Results of drug target analysis were uploaded (stable genes based on data by Hicks et al.,) Galaxy Tool Upload File Upload File Upload File Upload File Complement on data 4 Coverage on data 3 and data 5 Intersect on data 3 and data 5 UCSC Main Table Browser: refGene (genome) UCSC Main Table Browser: ccdsGene (genome) Upload File Upload File Upload File Join two Datasets on data 11 and data 12 Upload File Join two Datasets on data 10 and data 14 Upload File Upload File Join two Datasets on data 16 and data 17 Upload File Upload File Join two Datasets on data 19 and data 20 Upload File 23 24 Uploaded list of stable genes (stable in copy number as well as expression based on data by Chin and Naderi et al.,) Determined which stable genes were targets of drugs based on previous drug analysis (Hicks et al.,) Upload File Join two Datasets on data 22 and data 23