etc2514-sm-0001-SuppData
advertisement
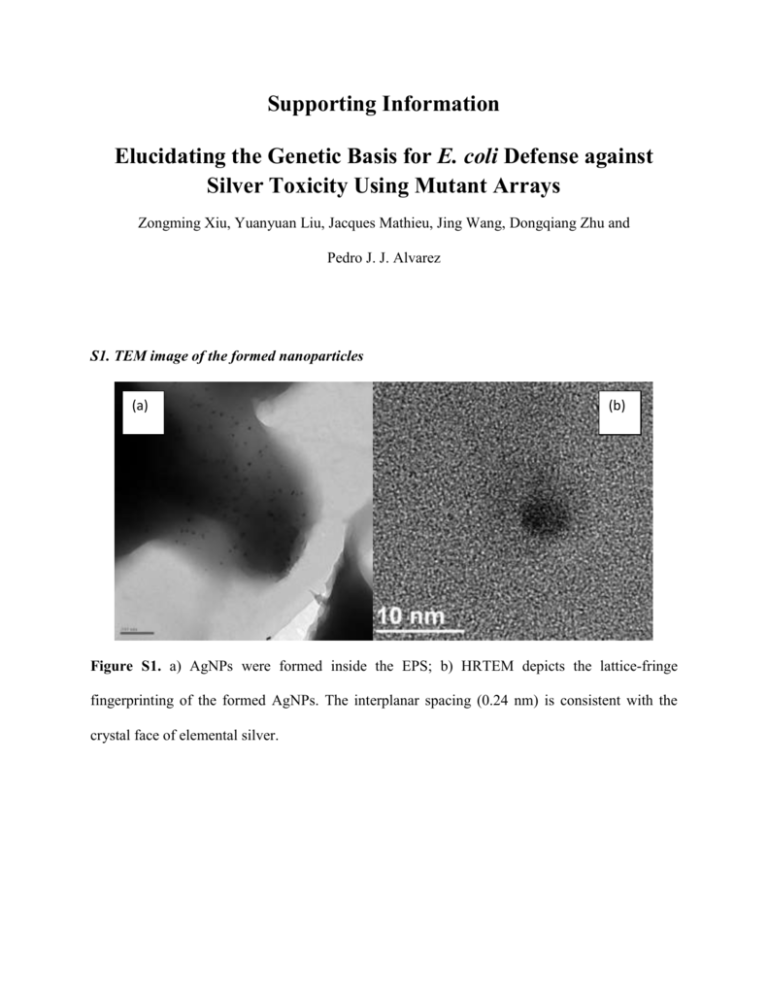
Supporting Information Elucidating the Genetic Basis for E. coli Defense against Silver Toxicity Using Mutant Arrays Zongming Xiu, Yuanyuan Liu, Jacques Mathieu, Jing Wang, Dongqiang Zhu and Pedro J. J. Alvarez S1. TEM image of the formed nanoparticles (a) (b) Figure S1. a) AgNPs were formed inside the EPS; b) HRTEM depicts the lattice-fringe fingerprinting of the formed AgNPs. The interplanar spacing (0.24 nm) is consistent with the crystal face of elemental silver. S2. TEM characterization of bacterial cells under silver stress (a) (b) Figure S2. Morphology of E. coli cells exposed to AgNO3 (8 mg/L). (a) The cytoplasm of the wild type E. coli contracted away from the cell envelope. (b) Cell debris without intact cells were observed in the pgaB culture. S3. Protein production by ORF clones Protein produced (OD278nm) 0.25 * 0.20 * 0.15 * 0.10 0.05 0.00 WT WT/BP sodB recA Figure S3. Higher amount of proteins are produced in the ORF clones that over-express specific genes (wild type with blank plasmid (WT/BP), sodB, recA) than the wild type control (WT), and these proteins (regardless of the gene that codes them) provide binding sites for Ag+, making ORF clones more resistant to silver. Asterisks (*) denote significantly higher values than WT control (p < 0.05). The protein concentration was measured by the Bradford method 1. Wild type E. coli and ORF mutants were grown overnight (16h), and diluted with DI water to the same OD600 value (0.8). Cells were washed and harvested by centrifugation (10,000 rpm) and lysed using B-PER Bacterial protein extraction reagent (Thermo Scientific) to dissolve proteins. The protein concentrations were measured using a spectrophotometer (Ultraspec 2100 pro, Amersham Biosciences Inc. NJ, USA) at OD 278nm. Reference 1. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1): 248-254.