Chemostat Gr1 rcr80
advertisement

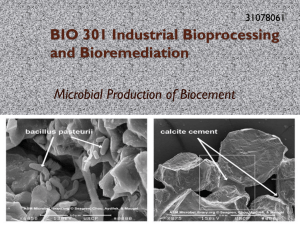
Murdoch University Chemostat Report Group 1 Rodney Ling – 32140372, Kristen Wolter – 31250128, King Zheng (Sam) Lim – 31544501 10/1/2013 Abstract Biocementation process has utilized the ability of microbes to promote precipitation of 𝐶𝑎𝐶𝑂3 . This in turn will be used on repairing lime stone and strengthening concrete. Moreover, such ability has been employed for applications such as preventing soil avalanching and compacting soil from reclaimed land. One of such bacterium(of what such? What the word such referring to?) is S.pasteurii. The aim of this experiment was to investigate the viability of S.pasteurii growing in a non-sterile Chemostat by providing harsh environment at pH 10 to prevent other types of microbial growth. Good In addition, various parameters such as varying hydraulic retention time (HRT) of the chemostat, urea concentration and yeast concentration in feed media have been carried out to improve the purity of culture and to improve urease production. This explained what you have done. Now, I hope you explain what you have found. it was found that by increasing the HRT, biomass productivity increased; however it did not increase the specific urease activity of culture. Furthermore, results also showed that by performing yeast extract increment in the media unclear (adding YE?), the purity of the culture decreased quite significantly, along with a sign of decreasing specific urease activity. Nonetheless, such alteration caused a rise in biomass concentration. Besides, increasing urea concentration in the media not only did not show an increase in purity of the culture, but also showed a lower growth and specific urease activity of S.pasteurii in the culture vessel. Lastly, the highest urease activity product obtained from the Chemostat has shown success in biocementation of silicon sand. Reasonable abstract. Could refer to pH or a causal relationship between conditions and effects on urease activity. Keywords: Sporosarcina pasteurii, urease, biocementation Introduction In almost every environment on Earth there are microbes which participate in mineralization processes such as calcium carbonate precipitation (Achal et al, 2009). In both laboratory and natural settings, microbes have shown to induce the precipitation of calcium carbonate and are thus regarded as important factors in the formation of carbonate sediments and deposits (Achal et al, 2009). Bioremediation processes have utilised this ability of microbes to create a microbial plugging process, known as biocementation, to promote the precipitation of calcium carbonate in the form of calcite (Achal et al, 2009). This is then used for various applications such as the consolidation of sand columns, the repair of limestone monuments and the repair of cracks in concrete (Achal et al, 2009). Furthermore, using biocementation to enhance the strength and stiffness of soil, soil avalanching can be prevented, soil on reclaimed land sites can be compacted, and the effects of liquefaction can be alleviated (Ivanov et al, 2008). One such microbe which has been shown to contribute to calcium carbonate precipitation is Sporosarcina pasteurii (previously known as Bacillus pasteurii), an endospore-forming, alkaliphilic, soil bacterium, which grows at an optimum pH of 9 (Achal et al, 2009). S.pasteurii produces a nickel containing enzyme called urease which catalyses the hydrolysis of urea to ammonia and carbon dioxide: 𝑁𝐻2 − 𝐶𝑂 − 𝑁𝐻2 + 𝐻2 𝑂 → 2𝑁𝐻4+ + 𝐶𝑂32− Nice and clear to here. This results in an increase in pH in the immediate environment, which when in the presence of calcium and carbonate ions causes them to precipitate together as calcium carbonate (Stocks-Fischer et al, 1999). Thus, by integration of S.pasteuriiwith urea and calcium chloride, biocementation can occur. It has also been found that the presence of high concentrations of ammonia does not inhibit growth of S.pasteurii, nor does it prevent urease production as it does in other microorganisms which produce urease such as Pseudomonas 1 aerogenes (Morsdorfet al, 1989). This combined with the high pH optimum of S.pasteurii, allows for selective growth conditions even in a non-sterile environment. Source? (Cheng paper?) From the equation above, the production of ammonium ion can result in ammonia gas formation. As a consequence, it can cause some degree of pH drop and affect the culture purity. Such reaction can be explained by using the Le’ Chatelier’s principle (Appendix G), with support of the spontaneity of the reaction using Gibbs free energy. However, it has also been found that the presence of high concentrations of ammonia does not inhibit growth of S.pasteurii, nor does it prevent urease production as it does in other microorganisms which produce urease such as Pseudomonas aerogenes (Morsdorf et al, 1989). This combined with the high pH optimum of S.pasteurii, allows for selective growth conditions even in a nonsterile environment. In this project, a chemostat was run for two weeks under non-sterile conditions containing a culture of S.pasteurii. A constant temperature of 280 Celsius and a pH of approximately 10 were used for the chemostat. The aim of the experiment was to test the feasibility of S.pasteurii production of urease under continuous conditions, in a non-sterile chemostat, employing harsh conditions to minimize contamination and to optimize conditions to enhance the production of urease. Nicely defined aim. This elevates the introducition to a more quality level. Various methods were employed to enhance the production of the enzyme including, altering the concentration of urea and yeast extract in the feed, changing the hydraulic retention time of the chemostat and controlling the pH of the environment to limit contamination. Following the two weeks of inoculation, a harvest sample with the highest specific urease activity was used to attempt biocementation to determine if production of urease in a non-sterile condition is feasible. I would better finish the intro with the aim rather than pre-empting results. Methods Chemostat Set Up: A 600ml bioreactor was placed in a 28oC water bath with an overhead drill stirrer, an airflow tube and feed and harvest tubes connected into it.The airflow tube was connected to an airflow meter which was connected to an air source. Both the feed and harvest tubes were connected to separate pumps, which had corresponding tubes connected to a feed bottle and a harvest bottle. The pumps were also connected through a labjack data card to a computer so that the chemostat system could be controlled via the computer. A diagrammatic representation of the chemostat set up can be seen in Figure 1. Note: as the pH pump was not working correctly from batch 1 result thus, it was disassembled from the set up for batch 2 culture. Could explain more how the pH was maintained. 2 Figure 1: Diagrammatic representation of chemostat set up. good Chemostat Innoculation and Running: 600ml of S.pasteurii culture was inoculated into the bioreactor at the beginning of the two weeks. The chemostat was continuously fed and harvested over the two weeks with retention times altered according to needs of the experiment. The harvest pump was set at the 600ml mark to enable a consistent volume of 600ml in the chemostat. The stirring speed was set at a constant 400rpm, and airflow rates were varied according to experiment needs. Temperature of the chemostat was kept at a constant 28oC, and pH was manually controlled to maintain an approximate pH of 10. Urease Activity: The urease activity of the culture was determined using a conductivity assay. This was achieved by adding10ml of 3M urea, 8ml of deionized water and 2ml of culture to a 50ml centrifugation tube. A conductivity probe was then placed into the tube and was used to gently mix the solution. Conductivity changes were then measured over 10 minutes at room temperature. The conductivity probe was calibrated prior to each test following instructions provided by the manufacturer. Could briefly explain the principle Chemostat Feed: 20g of yeast, 10.21g of urea, 20g of sodium acetate and 2ml of nickel chloride (50mM stock solution) were combined and deionized water added to make up to 900mls. The pH was then adjusted to 10 using 10M sodium hydroxide before deionized water was added to make up the total volume of 1L. Following this, any further pH adjustments were made to maintain a pH of 10. Alterations in feed media were made by changing the amount of yeast or urea added. Dissolved oxygen, Optical Density and pH: Dissolved oxygen in the chemostat was measured using a DO probe. The probe was calibrated before every use following manufacturer instructions and measurements made in ppm. Optical density measurements were made using a spectrophotometer with a wavelength of 600nm, and using feed solution as the blank. To obtain a reading under 1, various dilutions were made and the resultant reading multiplied by the dilution factor to determine true optical density. The pH was also measured using a pH probe. The probe was calibrated prior to each use following manufacturer instructions and the pH adjusted up to 10 using 10M sodium hydroxide. 3 Biocementation: Based on the production of carbonate ions from urease activity, biocementation was performed by adding 10ml of harvest culture with a urease activity of 31 mM/min into a 50ml syringe filled with silicon sand. After the excess harvest culture had been flashed out via rubber tube, 10ml of 1M calcium chloride and 1M of urea was added. Subsequently, 20ml of 1M calcium chloride and 1M of urea was added every 7 hours. By providing calcium ions and urea to S.pasteurii, formation of carbonate ions can be formed by urease activity and precipitation of calcium carbonate to the sand would occur. Figure 2. Sand Column set up. 4 good methods section Results and Discussions Starting up of chemostat in a non-sterile condition (Batch 1) During the first week of chemostat project, various experimental parameters were determined prior to start up, in order to achieve a successful running chemostat. Parameters such as the retention time and feed media selection were considered and were varied accordingly. For 24 hours of retention time operation, an initial optical density (OD) of the culture in the bioreactor was read at 0.585 absorbance (abs). Based on Liang’s findings for the conversion of cell turbidity to culture biomass concentration (Cheng & Cord-Ruwisch, 2013), the biomass concentration was found to be 0.2574 g/L. show calculation, this value sounds too low to me. In the meantime, an initial urease activity test was carried out and its activity was found to be 4.889 mM/min. Thus, initial specific urease activity was determined to be 8.3325 mM/min/OD (Refer to Appendix B, Batch 1, Day 0). An issue with the harvest pump resulted in a slight delay, with time taken to trouble shoot required, and the discovery that the harvest pump was unserviceable. Consequently, this fault caused the culture media in the bioreactor to be diluted, thus giving a low optical density reading. Therefore, the logical decision was made to increase the retention time to 48 hours in order to increase the biomass. Could go in appendix. The reader is interested in how you test your idea of producing more of the enzyme (hypothesis / scientific method) Optical density was determined to be 0.828 abs after the change in hydraulic retention time. This relates to a higher biomass concentration which was found to be at 0.364g/L. Subsequently, its specific urease activity was found to be 21.8867 mM/min/OD (Refer to Appendix B, Batch 1, Day 1). Such alteration on the HRT had significantly increased the biomass concentration and its specific urease activity, which suggested that the culture growth and urease production had increased to its steady state. High NaOH consumption due to pH meter inaccuracy An unexpected event, involving an error related to the pH Pump, resulted in an additional 50ml of 10mM NaOH being added overnight. A re-calibration was then necessary in order to bring the system back to the required state. At the same time, due to the jumpiness of the raw data measured by the pH probe, some improvement on filtering the unwanted raw data was done by implementing an error range filtration in the Chemostat program, within Labview. After debugging the program, the measured pH of the culture was seen to drastically reduce to 6.58. Due to the fact the apparent pH value was below the optimum (pH10), 5M of NaOH was added continuously by running the NaOH pump for approximately 15 minutes. After a further measurement revealed the pH had overshot to 14, it was again adjusted accordingly with the addition of 150ml of 5M HCl. This action was effective in raising the pH to the acceptable level of 10.24. Fair enough, the exposure to pH 14 could explain loss of the culture , which would be much nicer shown as time course figure than a table. The chemostat was left for 48 hrs to sequence a batch culture, with the airflow rate set at 100 L/h, and stirrer at 200 rpm. Later when checked, the bioreactor was found overflowing with a frothing culture that was sticky and pale-white in colour. This gooey-textured growth, found swarming around the reactor as well as the water bath, was not S.pasteurii according to its morphology. The reactor was therefore confirmed to be contaminated and the chemostat was re-inoculated with new culture. Due to the issues involving pH and high consumption of NaOH overnight, it was derived that the NaOH pump was faulty (it remained in operation and pumped NaOH in disregard of the pH point set in the Labview program). Consequently, the pH level was controlled manually for the remainder of the experiment. 5 New inoculant of culture (Batch 2) For the new inoculant added into the bioreactor, its initial OD reading was 1.48 abs, which lead to a biomass concentration 0.651g/L and specific urease activity 11.0274 mM/min/OD. The culture was fed with a 2 fold increment in yeast extract to improve growth, and a HRT of 48 hours. This setting was run for the first 24 hours to increase the biomass for subsequent experiments, and this was set as Day 0 of the chemostat operation. The urease activity, OD reading as well as the biomass concentration for each day was successfully recorded and is tabulated below: Table 1. Successive OD reading, biomass concentration and urease activity on daily basis. (Appendix B, Batch 2) biomass concentration g/L urease activity mS/min urease activity mM/min specific urease activity mM/min/OD 1.480 0.6512 0.1469 16.3205 11.0274 - - - 5.283 2.3245 0.2218 24.6419 4.6644 12.5 0.0208 0.0484 11.700 5.1480 0.4099 45.5398 3.8923 25 0.0417 0.2145 9.280 4.0832 0.3319 36.8740 3.9735 50 0.0833 0.3403 9.290 4.0876 0.2393 26.5862 2.8618 50 0.0833 0.3406 7.500 3.3000 0.1160 12.8876 1.7184 - - - 5.000 2.2000 0.1075 11.9432 2.3887 50 0.0833 0.1833 OD day 0 day 1 day 2 day 3 day 4 day 7 day 8 2x yeast extract 2x urea 2x yeast extract flow rate mL/hr dilution rate h-1 productivity gX/L/h Overall good recording of data. It is always more instructive to plot some data to show time courses and effects of changes as a function of time. Arguably the key production value is the enzyme productivity, which was not recorded. Only use significant (meaningful) decimal places ! Giving 4 decimal places implies that you can determine values to that precision and that it is important for the interpretation of trends to show them. So clearly less is more. It can be seen that after 1day (Table 1, Day 1) the biomass increased with an OD reading of 5.2 abs. This lead to an increase in biomass concentration of 2.3g/L with a specific ureases activity reading at 4.6 mM/min/OD, while the pH of the culture dropped to 9.3 during the operation. Due to such low readings (which? Be specific) compared to the optimum pH, the system was not harsh enough to eliminate other microbes growth, thus a manual increment on pH was performed using 10mM NaOH solution to bring up the reactor pH to 10. A drop in specific urease activity might signify that the bacterial culture was less pure than of the initial inoculum on the previous day. After running for a day with a dilution rate of 0.04167 (Table 1, Day 2) and comparing to Day 1’s dilution rate 0.0208 hr-1, it was noted that an approximate 2 fold increment in the biomass concentration plus an increase in urease activity by almost 2 fold has occurred. Nonetheless, a drop in specific urease activity can be seen from 4.6644 to 3.8923 mM/min/OD, which implies that the culture purity was relatively low, compared to that of the previous day. Thus, a trial run on increasing the dilution rate was tested to determine if an increase in dilution rate would increase the purity of a culture in conditions as harsh as pH 10. Effects of HRT on chemostat It was found that increasing the dilution rate lowered the biomass concentration from 5.148g/L to 4.0832g/L (Table 1, Day 2). Urease activity has been seen to drop from 45.5398 mM/min to 36.874 mM/min but its specific urease activity was about the same as Day 2 (3.8923 mM/min/OD). Therefore, it can be stated that 6 an increase in dilution rate would not affect the urease activity; however it would increase its biomass productivity from 0.2145gX/L/h to 0.34gX/L/h, provided that the dilution rate was not at its critical point. Next, a trial on increasing urea content in the media was done to determine the significance of urea concentration on the purity of culture, and to determine the effect of high production of NH3 on culture growth. Effects of urea concentration on purity of culture Based on the result for Day 4, due to the same dilution, the biomass concentration and productivity remained the same. On the other hand, a decrease in urease activity and specific urease activity was seen in the culture. Theoretically, this trial should have increased the purity of the culture and it would have increased in urease activity and specific urease activity should be significant. Nevertheless, due to a decline in pH at 9.52 after 24 hours operation, there can be a possibility that such pH was not at its optimum, hence causing a decline in urease activity (Cheng & Cord-Ruwisch, 2013). Effect of batch culture Chemostat was turned off during the weekends and was found to have 100ml of culture left after 48 hours. Evaporation could have been one of the factors that caused this problem as it was noticed that the lab window adjacent to the lab bench was left open throughout the weekend. Similarly, the water level in water bath was noticeably low too. Another factor that may give arise to this problem is that since a large amount of ammonia was produced before turning off the chemostat, there could be a possibility of diffusion to occur due to the volatility of ammonia. The equation below shows the equilibrium reaction when ammonium ion interacts with strong base. 𝑁𝐻4+ (𝑎𝑞) + 𝐻2 𝑂(𝑙) ↔ 𝑁𝐻3 (𝑔) + 𝐻3 𝑂+ (𝑎𝑞) Or, 𝑁𝐻4+ (𝑎𝑞) + 𝑂𝐻 − (𝑎𝑞) ↔ 𝑁𝐻3 (𝑔) + 𝐻2 𝑂(𝑙) By referring Le’ Chatelier’s principle (refer to Appendix G), in high pH condition, since hydroxide ions (𝑂𝐻 − ) are abundant, equilibrium will shift to the right, hence increases the formation of ammonia gas. At the same time, since hydroxide ions are being used to form water molecule, from the second equation, protons will be gradually increased over time. As a result of that, the reactor pH will be slightly decreased due to the reduction of protons in the culture and hence both urease activity and the biomass concentration declined quite significantly. To support that, Standard Gibbs free energy of formation is used to explain the spontaneity of the formation of ammonia gas. Assuming that the ambient temperature for the above reaction was 25℃ and the surrounding reactor vessel was at 1 atm pressure, the Gibbs free energy of the above reaction is ∴ ∆𝐺° = −17.03 𝑘𝐽 𝑚𝑜𝑙 −1 Since the Gibbs free energy has a negative value, this signifies that the above reaction is spontaneous in forward reaction, thus favoring the formation of ammonia gas. Thus, this supports the claim that a drop in pH reactor vessel was responsible to the ammonia gas formation, and consequently causing a dramatic volume loss in the reactor during the batch culture sequencing. Optimizing biomass concentration and urease production for handing over At day 7, the chemostat was kept running with twice the amount of yeast extract in the feed media. This in turn increased the biomass and the urease activity of the culture in order to let the next group taking over the chemostat. Based on the observation from day 7 and day 8, its specific urease activity had increased, but the biomass concentration dropped significantly (Table 1). One probable reason may due to an accumulation of ammonia concentration in the bioreactor during the period of shut down. As a consequence of that, an 7 inhibition on bacterial growth may have occurred and this might lead to cell lysis. Therefore, this might cause a decrease in urease activity. Apart from that, pH might be part of the reason why the urease activity was lowered over the period of shut down since the pH in the reactor was around 9.5. It would have been nice to show a time course of performance data such as biomass productivity, urease activity, specific activity etc. Also you did not not use a data processing spreadsheet as in bioprosim. 8 Biocementation Product Figure 3. Biocementation Product. Biocementation was done with the highest specific urease activity of 31mM/min. After 2 weeks of biocementing, it was observed that the loose silicon sand found in the syringe has compacted together and formed a rock like structure (Figure 3). Due to slow rate of biocementation and time constraint, biocementation was not complete thus a full sand column cementation was not established. In conclusion, based on the biocementation result, biocementation using S. pasteurii is feasible as cementation was a success. OK Possible improvements Biocementation should be done with a controlled pump (probably by implementing a Labview program), allowing a continuous input of calcium chloride and urea every 7 hours. AS a result, a better production of sand column can be achieved. Pumps should be checked before allowing student to temper as to avoid confusion among students if nonworkable equipment were given. A short briefing regarding tasks to be done in chemostat before the lab should be provided. This allowed students to do some research on the bacteria and the setting up of the chemostat prior to the day of the experiment. Conclusion Chemostat was set-up and found that it is feasible to grow in an S.pasteurii swap words non-sterile environment. Using “harsh” conditions, we could minimize the contamination to occur by allowing pH to be set at 10. increasing of yeast extract in the media increased the biomass concentration of culture but also the contaminant level as could be seen from a lower specific urease activity. A change in HRT did not have effect on the purity of culture as specific urease activity remained the same but with an increase of HRT, it incurred an increase in productivity. Increasing of Urea concentration in the media did not showed an increase of purity of culture but showing a lower growth of S.pasteurii and lowering of specific urease activity. Since the parameter was unstable during the occurrence of experiment, it showed an inconclusive result. The product of Chemostat has shown success in biocementation of silicon sand. This is more a summary rather than conclusion (what do the results mean for example for industry). 9 References Achal, V., Mukherjee, A., Basu, P. and Sudhakara Reddy, M. (2009). “Strain improvement of Sporosarcina pasteurii for enhanced urease and calcite production”. Journal of Industrial Microbiology and Biotechnology. 36.7:981-988. Atkins, P.; Paula, J.d. (2010). Atkins' Physical Chemistry, 9th ed. Oxford; New York: Oxford University Press, 2010; pp922-924. Cheng, L. and Cord-Ruwisch, R. (2013). Selective enrichment and production of highly urease active bacteria by non-sterile (open) chemostate culture. Society for Industrial Microbiology and Biotechnology. Ivanov, V. and Chu, J. (2008).“Applications of microorganisms to geotechnical engineering for bioclogging and biocementation of soil in situ”.Reviews in Environmental Science and Bio/Technology. 7: 139-153. Stocks-Fischer, S., Galinat, J and Bang, S. (1999). “Microbiological precipitation of CaCO3”.Soil Biology and Biochemistry. 31:1563-1571. Victoria, S. W. (2004). Microbial 𝐶𝑎𝐶𝑂3 Precipitation for the Production of Biocement. Perth, Western Australia: School of Biological Sciences & Biotechnology, Murdoch University. Don’t use people’s first names in citations instead of their family name 10 Appendices Appendix A: Hydraulic Retention Time (HRT) calculation (Pump ON and Pump OFF ratio) Given that the bioreactor volume was 600mL and the retention time was set to be 24 hour, 𝑖𝑒. 𝑅𝑒𝑡𝑒𝑛𝑡𝑖𝑜𝑛 𝑇𝑖𝑚𝑒 = 600𝑚𝐿/24ℎ𝑟 = 25𝑚𝐿/ℎ𝑟 = 0.416𝑚𝐿/𝑚𝑖𝑛 The feed pump flow rate was determined to be 11.5mL/min, by working out the ratio of the feed pump flow rate to the retention time required, 𝐻𝑅𝑇 → 𝑓𝑒𝑒𝑑 𝑝𝑢𝑚𝑝: 𝑅𝑒𝑡𝑒𝑛𝑡𝑖𝑜𝑛 𝑡𝑖𝑚𝑒 𝑖𝑒. 11.5 𝑚𝐿 𝑚𝐿 ∶ 0.416 𝑚𝑖𝑛 𝑚𝑖𝑛 ∴ 27.6 ≈ 28 𝑚𝑖𝑛 ∶ 1 𝑚𝑖𝑛 The ratio determined above signifies that the feed pump will be turned on for 1 minute, and then followed by switching off the feed pump for about 28 minutes. Appendix B: Chemostat Raw Data and Yield Coefficient Calculation Batch 1: Urease activity (mathematical) Urease activity (graphical) OD specific activity yield coefficient Conductivity as a function of time Conductivity(ms) 4.4 4.4 4.43 4.53 4.56 4.63 4.69 4.73 4.76 4.76 4.79 0.039 0.044 0.585 0.075213675 0.006435 4.9 4.85 4.8 4.75 4.7 4.65 4.6 4.55 4.5 4.45 4.4 4.35 y = 0.0446x + 4.3841 R² = 0.9612 Conductivity (ms) Day 0 - 5/09/2013 Time (min) 0 1 2 3 4 5 6 7 8 9 10 Series1 Linear (Series1) 0 5 10 time (min) ms/min ms/min abs ms/OD/min gX/gS Media Content – 20g Yeast Extract + 20g Sodium acetate + 10.21g Urea Substrate Content – 40g 11 Urease activity (mathematical) Urease activity (graphical) OD specific activity yield coefficient Conductivity(ms) 3.82 4.66 4.85 5.23 5.38 5.56 5.63 5.7 5.78 5.88 5.92 5.96 0.214 0.163 0.828 0.196859903 0.009108 Conductivity as a function of time Conductivity (ms) Day 1 - 6/09/2013 Time (min) 0 0.5 1 2 3 4 5 6 7 8 9 10 y = 0.1636x + 4.6073 R² = 0.7672 7 6 5 4 3 2 1 0 Series1 Linear (Series1) 0 5 10 time (min) ms/min ms/min abs ms/OD/min gX/gS Media Content – 20g Yeast Extract + 20g Sodium acetate + 10.21g Urea Substrate Content – 20g Batch 2: Urease activity (mathematical) Urease activity (graphical) OD specific activity Conductivity(ms) 6.15 6.28 6.38 6.68 6.88 6.96 7.13 7.25 7.35 7.43 7.55 7.67 0.152 0.1469 1.48 0.099256757 Conductivity as a function of time Conductivity (ms) Day 0 – 9/9/2013 Fresh Culture Time (min) 0 0.5 1 2 3 4 5 6 7 8 9 10 9 8 7 6 5 4 3 2 1 0 y = 0.1469x + 6.2962 R² = 0.9656 Series1 Linear (Series1) 0 5 10 time (min) ms/min ms/min abs ms/OD/min Media Content – 40g Yeast Extract + 20g Sodium acetate + 10.21g Urea Substrate Content – 60g 12 Urease activity (mathematical) Urease activity (graphical) OD specific activity yield coefficient Conductivity(ms) 5.94 6.01 6.07 6.3 6.53 6.74 6.94 7.15 7.29 7.71 7.9 8.15 0.221 0.2218 5.283 0.041983721 0.038742 Conductivity as a function of time Conductivity (ms) Day 1 - 9/10/2013 Time (min) 0 0.5 1 2 3 4 5 6 7 8 9 10 9 8 7 6 5 4 3 2 1 0 y = 0.2218x + 5.8686 R² = 0.9941 Series1 Linear (Series1) 0 5 10 time (min) ms/min ms/min abs ms/OD/min gX/gS Media Content – 40g Yeast Extract + 20g Sodium acetate + 10.21g Urea Substrate Content – 60g Urease activity (mathematical) Urease activity (graphical) OD specific activity yield coefficient Conductivity(ms) 5.6 5.8 6.01 6.44 6.85 7.25 7.7 8.05 8.44 8.87 9.29 9.73 0.413 0.4099 11.7 0.035034188 0.1287 Conductivity as a function of time 12 Conductivity (ms) Day 2 - 9/11/2013 Time (min) 0 0.5 1 2 3 4 5 6 7 8 9 10 y = 0.4099x + 5.6066 R² = 0.9998 10 8 Series1 6 Linear (Series1) 4 2 0 0 5 10 time (min) ms/min ms/min abs ms/OD/min gX/gS Media Content – 20g Yeast Extract + 20g Sodium acetate + 10.21g Urea Substrate Content – 40g 13 Urease activity (mathematical) Urease activity (graphical) OD specific activity yield coefficient Conductivity(ms) 5.12 5.4 5.56 5.99 6.31 6.62 6.94 7.27 7.57 7.88 8.2 8.53 0.341 0.3319 9.28 0.035765086 0.10208 Conductivity as a function of time Conductivity (ms) Day 3 - 9/12/2013 Time (min) 0 0.5 1 2 3 4 5 6 7 8 9 10 y = 0.3319x + 5.2474 R² = 0.9976 9 8 7 6 5 4 3 2 1 0 Series1 Linear (Series1) 0 5 10 time (min) ms/min ms/min abs ms/OD/min gX/gS Media Content – 20g Yeast Extract + 20g Sodium acetate + 10.21g Urea Substrate Content – 40g Urease activity (mathematical) Urease activity (graphical) OD specific activity yield coefficient Conductivity(ms) 6.02 6.12 6.26 6.54 6.76 6.99 7.23 7.46 7.7 7.93 8.18 8.42 0.24 0.2393 9.29 0.025758881 0.10219 Conductivity as a function of time Conductivity (ms) Day 4 - 9/13/2013 Time (min) 0 0.5 1 2 3 4 5 6 7 8 9 10 9 8 7 6 5 4 3 2 1 0 y = 0.2393x + 6.0274 R² = 0.9997 Series1 Linear (Series1) 0 5 10 time (min) ms/min ms/min abs ms/OD/min gX/gS Media Content – 20g Yeast Extract + 20g Sodium acetate + 20.42g Urea Substrate Content – 40g 14 Urease activity (mathematical) Urease activity (graphical) OD specific activity yield coefficient Conductivity(ms) 7.51 7.61 7.68 7.78 7.94 8.02 8.13 8.26 8.39 8.47 8.58 8.7 0.119 0.116 7.5 0.015466667 0.0825 Conductivity as a function of time Conductivity (ms) Day 7 - 9/16/2013 Time (min) 0 0.5 1 2 3 4 5 6 7 8 9 10 y = 0.116x + 7.5526 R² = 0.9972 8.8 8.6 8.4 8.2 8 7.8 7.6 7.4 Series1 Linear (Series1) 0 5 10 time (min) ms/min ms/min abs ms/OD/min gX/gS Media Content – 20g Yeast Extract + 20g Sodium acetate + 20.42g Urea Substrate Content – 40g Urease activity (mathematical) Urease activity (graphical) OD specific activity yield coefficient Conductivity(ms) 7.17 7.12 7.11 7.31 7.46 7.56 7.67 7.78 7.87 7.95 8.06 8.15 0.098 0.1075 5 0.0215 0.03667 Conductivity as a function of time Conductivity (ms) Day 8 - 9/17/2013 Time (min) 0 0.5 1 2 3 4 5 6 7 8 9 10 8.4 8.2 8 7.8 7.6 7.4 7.2 7 y = 0.1075x + 7.1035 R² = 0.986 Series1 Linear (Series1) 0 5 10 time (min) ms/min ms/min abs ms/OD/min gX/gS Media Content – 40g Yeast Extract + 20g Sodium acetate + 10.21g Urea Substrate Content – 60g 15 Appendix C: Biomass Measurement (𝑂𝐷600 ) According to Cheng and Cord-Ruwisch’s finding (Cheng & Cord-Ruwisch, 2013), due to a good correlation between biomass concentration and optical density (OD), a relationship was established and it can be seen as below: 𝐵𝑖𝑜𝑚𝑎𝑠𝑠 𝑐𝑜𝑛𝑐. (𝑔/𝐿) = 0.44 × 𝑂𝐷(600 𝑛𝑚) Hence, by determining the optical density of the reactor daily, its biomass concentration can be calculated for further calculation. Noted that the optical density used for the calculation was the neat reading, ie. Not the dilution reading from spectrophotometer. Appendix D: Yield coefficient calculation To work out the yield coefficient for the culture in the bioreactor, the amount of substrate used in the feed is needed, as well as the biomass concentration in the culture of that day. By using the relationship below: 𝑌𝑖𝑒𝑙𝑑 𝐶𝑜𝑒𝑓𝑓𝑖𝑐𝑖𝑒𝑛𝑡(𝑔𝑋/𝑔𝑆) = 𝐵𝑖𝑜𝑚𝑎𝑠𝑠 𝑐𝑜𝑛𝑐. (𝑔/𝐿) 𝑆𝑢𝑏𝑠𝑡𝑟𝑎𝑡𝑒 𝑐𝑜𝑛𝑐. (𝑔/𝐿) Appendix E: Urease activity and Specific urease activity calculation According to the Appendix A, the daily urease activity was calculated using the expression below: 𝑈𝑟𝑒𝑎𝑠𝑒 𝐴𝑐𝑡𝑖𝑣𝑖𝑡𝑦 (𝑚𝑠/𝑚𝑖𝑛) = 𝑈𝑓 − 𝑈𝑖 𝑡𝑓 − 𝑡𝑖 Where, 𝑈𝑓 and 𝑈𝑖 is the conductivity of the sample at the end of the time interval and at the beginning of the time interval respectively, 𝑡𝑓 − 𝑡𝑖 is the time interval. As reported by Victoria (Victoria, 2004) and as stated in Cheng’s finding (Cheng & Cord-Ruwisch, 2013), the hydrolyzed urea from the conductivity test (in mS) can be converted to the amount of ammonium produced (in mM) at the end of hydrolysis, and it can be shown as the following expression: 𝑈𝑟𝑒𝑎𝑠𝑒 𝑎𝑐𝑡𝑖𝑣𝑖𝑡𝑦 (𝑚𝑀/𝑚𝑖𝑛) = 𝐶𝑜𝑛𝑑𝑢𝑐𝑡𝑖𝑣𝑖𝑡𝑦 (𝑚𝑆/𝑚𝑖𝑛) × 111.1 Thus, the amount of urease in the bioreactor for each daily can be calculated. Apart from that, the specific urease activity in relation to the biomass of the culture can be determined using the below relationship: 𝑆𝑝𝑒𝑐𝑖𝑓𝑖𝑐 𝑢𝑟𝑒𝑎𝑠𝑒 𝑎𝑐𝑡𝑖𝑣𝑖𝑡𝑦 (𝑚𝑀/𝑚𝑖𝑛/𝑂𝐷) = 𝑈𝑟𝑒𝑎𝑠𝑒 𝑎𝑐𝑡𝑖𝑣𝑖𝑡𝑦 (𝑚𝑆/𝑚𝑖𝑛) 𝑂𝐷 Appendix F: Feed pump flow rate, reactor dilution rate and biomass productivity calculation By setting the reactor volume as 600mL and changing the retention time, the dilution rate of the reactor can be determined by using the following expressions. 𝐹𝑙𝑜𝑤 𝑟𝑎𝑡𝑒 (𝑚𝐿/ℎ𝑟) = 𝐷𝑖𝑙𝑢𝑡𝑖𝑜𝑛 𝑅𝑎𝑡𝑒 (ℎ𝑟 −1 ) = 600𝑚𝐿 𝑅𝑒𝑡𝑒𝑛𝑡𝑖𝑜𝑛 𝑡𝑖𝑚𝑒 (ℎ𝑟) 𝐹𝑙𝑜𝑤 𝑟𝑎𝑡𝑒 (𝑚𝐿/ℎ𝑟) 𝑅𝑒𝑎𝑐𝑡𝑜𝑟 𝑉𝑜𝑙𝑢𝑚𝑒 (𝑚𝐿) Similarly, the biomass productivity can be calculated using the expression below: 𝐵𝑖𝑜𝑚𝑎𝑠𝑠 𝑃𝑟𝑜𝑑𝑢𝑐𝑡𝑖𝑣𝑖𝑡𝑦(𝑔𝑋/𝐿/ℎ𝑟) = 𝐷𝑖𝑙𝑢𝑡𝑖𝑜𝑛 𝑅𝑎𝑡𝑒 (ℎ𝑟 −1 ) × 𝐵𝑖𝑜𝑚𝑎𝑠𝑠 𝑐𝑜𝑛𝑐. (𝑔/𝐿) By summarizing the calculated quantities and tabulated it, all of the results can be shown on Table 3. 16 Table 3. Feed Pump Rate, Dilution Rate and Productivity on Week 2 Chemostat Operation. day 0 day 1 day 2 day 3 day 4 day 7 day 8 Biomass Concentration (g/L) 0.6512 2.3245 5.1480 4.0832 4.0876 3.3000 2.2000 Retention time (hr) 48 24 12 12 12 Flow Rate (mL/hr) 12.5 25.0 50.0 50.0 50.0 Dilution Rate (h-1) 0.0208 0.0417 0.0833 0.0833 0.0833 Biomass Productivity (gX/L/h) 0.0484 0.2145 0.3403 0.3406 0.1833 Appendix G: Le’ Chatelier’s principle and Gibbs free energy calculation Ammonium ion, being slightly acidic in nature, will react with strong base to form a water molecule. Because its acid dissociation constant, 𝑝𝑘𝑎 value is 9.25, it is considered as a week acid. Therefore, acid-base reaction will take place. The equation below shows the equilibrium reaction when ammonium ion interacts with strong base. 𝑁𝐻4+ (𝑎𝑞) + 𝐻2 𝑂(𝑙) ↔ 𝑁𝐻3 (𝑔) + 𝐻3 𝑂+ (𝑎𝑞) Or, 𝑁𝐻4+ (𝑎𝑞) + 𝑂𝐻 − (𝑎𝑞) ↔ 𝑁𝐻3 (𝑔) + 𝐻2 𝑂(𝑙) Clearly, from the equation above, if the pH of the surrounding solution is low, according to Le’ Chatelier’s principle, equilibrium will shift to the left as protons (𝐻3 𝑂+ ) are abundant. Likewise, equilibrium will shift to the right if the pH is high as protons are limited. Due to the fact that S. pasteurii requires high pH environment for survival and growth, it is certain that some amount of ammonium ion will be converted to ammonia gas and causing a decrement on pH level. Certainly, the above equations are only true when the reaction is taken place in a close system. However, the above reaction can be applied to one of the reactions took place in the reactor vessel in order to establish further interpretation. From the table, Thermodynamic data for elements and inorganic compounds at 298K, (Atkins, 2010), the standard free energy of formation of each element are extracted and tabulated as below: Table 2. Thermodynamic data for elements and inorganic compounds at 298K. Elements ∆𝑓 𝐺° (𝑘𝐽 𝑚𝑜𝑙 −1 ) 𝑁𝐻3 (𝑔) -16.45 𝐻2 𝑂(𝑙) -237.13 𝑁𝐻4+ (𝑎𝑞) -79.31 𝑂𝐻 − (𝑎𝑞) -157.24 By using the following expression, Gibbs free energy is calculated, based on the second equation. ∆𝐺° = ∑ ∆𝑓 𝐺°𝑝𝑟𝑜𝑑𝑢𝑐𝑡 − ∑ ∆𝑓 𝐺°𝑟𝑒𝑎𝑐𝑡𝑎𝑛𝑡 ∆𝐺° = ((−16.45) + (−237.13)) 𝑘𝐽 𝑚𝑜𝑙 −1 − ((−79.31) + (−157.24)) 𝑘𝐽 𝑚𝑜𝑙 −1 ∴ ∆𝐺° = −17.03 𝑘𝐽 𝑚𝑜𝑙 −1 17 Appendix H: Hand over Notes The enzyme test and OD readings Make sure the conductivity probe is calibrated before using. After input of 10ml of urea 8ml of deionised water and 2ml of culture, remember to swirl the tube before starting the conductivity assay for accuracy Make sure no bubbles are in the probe by gentle taping to the bottom or stirring the solution using the probe. Have a constant time taken e.g. every min for 10 mins and set it for the rest of the conductivity test Always determine the gradient of conductivity against time. OD reading OD reading to be done with spectrophotometer at 600nm Using media which is your feed to blank the spectrophotometer and check the O.D of your sample *note mostly we did was 10x dilution for sample. You could do it for your samples. *note take readings lesser then 1 for accuracy -> based on Ralf Always calculate your specific activity which is gradient of slope / OD which gives u ms/OD/min to determine your activity for your findings and to determine your next move. Operate the chemostat Calculation of retention time and setting your bioreactor volume allowing it to be at a constant volume is critical. Determine of retention time based on feed in and off feed timing in relationship to the pump. Make an output (harvest) at a mark of your known volume and input will be feed in. Output pump rate must be higher than input to prevent overflowing. Remember to add antifoam solution (1 drop) if bioreactor start forming up *note anti-form solution is toxic to cells thus 1 drop will do it. You must have control on the inflow and outflow rate to determine your dilution rate. Test out the Noah micro pump and monitor closely on your Noah solution. Check water bath temperature and water level. Top up water if needed. Check D.O. in your reactor using probe. Run the computer for pH control In your group, we have a jumpy pH reading as compared to other groups. It is recommended that before connecting the NaOH pump, determine the lowest pH recorded from the probe in a pH10 buffer, and use that value as the pH setpoint on the NaOH pump. Remember to save the data of pH value and try running at 0.5 pumps in for NaOH and 120sec off. Due to the ‘jumpiness’ of the pH readings, a set of additional programming was done to filter out unwanted data points from the pH meter. You may not need to fully understand how the program works, but sufficient understanding on how to use it may aid in your operation. The red data points shown on the graph are the filtered data points, while the white data points show the raw data points from the pH probe. 18 If the pH probe picks up a big change in pH, the red data points will shift away from the raw data points. To overcome that, one should click on the ‘Filter’ button for a few moments. This allows the program to refresh itself and ‘readjust’ the filtered data points close to the interested data points. Also, noticed that the filter size will change to a value of 1000. Once the filtered data points are close to the interested data points unclick the button and change the filter size to 0.05. *note => always record your finding and your reasoning in the book and the next step you took for others to know what happened on that day to allow other team members to know what happened. 19