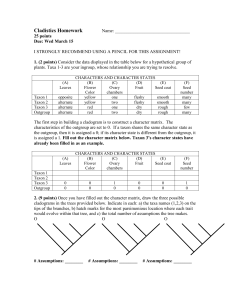
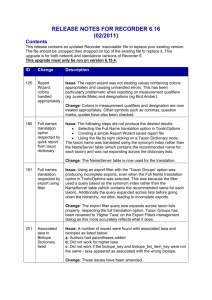
P A U P * Version 4.0a126 for Macintosh (X86) Thursday, February
advertisement

P A U P * Version 4.0a126 for Macintosh (X86) Thursday, February 21, 2013 12:05:43 PM Central Standard Time Running on IA-32 architecture (64-bit word length) SSE vectorization enabled Multithreading enabled for likelihood using Pthreads Compiled using Intel compiler (icc) 11.1.0 (build 20091012) -----------------------------NOTICE----------------------------This is an alpha-test version prepared for the exclusive use of course and workshop participants, as well as other authorized testers. It will expire on 1 Aug 2013. Please report bugs to david.swofford@duke.edu ---------------------------------------------------------------Processing of file "Seb Modified 21 Feb.nex" begins... Data matrix has 27 taxa, 53 characters Valid character-state symbols: 01234 Missing data identified by '?' Gaps identified by '-' Outgroup status changed: 3 taxa transferred to outgroup Total number of taxa now in outgroup = 3 Number of ingroup taxa = 24 Heuristic search settings: Optimality criterion = parsimony Character-status summary: Of 53 total characters: All characters are of type 'unord' All characters have equal weight All characters are parsimony-informative Gaps are treated as "missing" Multistate taxa interpreted as uncertainty Starting tree(s) obtained via stepwise addition Addition sequence: simple (reference taxon = Antarctican taxon) Number of trees held at each step = 1 Branch-swapping algorithm: tree-bisection-reconnection (TBR) with reconnection limit = 8 Steepest descent option not in effect Initial 'Maxtrees' setting = 100 Branches collapsed (creating polytomies) if maximum branch length is zero 'MulTrees' option in effect No topological constraints in effect Trees are unrooted Heuristic search completed Total number of rearrangements tried = 48442 Score of best tree(s) found = 146 Number of trees retained = 2 Time used = 0.01 sec (CPU time = 0.01 sec) Character types changed: Of 53 total characters: 1 character is of type 'ord' (Wagner) 52 characters are of type 'unord' *** Skipping "MESQUITECHARMODELS" block *** Skipping "MESQUITE" block Processing of input file "Seb Modified 21 Feb.nex" completed. paup> HSearch; Heuristic search settings: Optimality criterion = parsimony Character-status summary: Of 53 total characters: 1 character is of type 'ord' (Wagner) 52 characters are of type 'unord' All characters have equal weight All characters are parsimony-informative Gaps are treated as "missing" Multistate taxa interpreted as uncertainty Starting tree(s) obtained via stepwise addition Addition sequence: simple (reference taxon = Antarctican taxon) Number of trees held at each step = 1 Branch-swapping algorithm: tree-bisection-reconnection (TBR) with reconnection limit = 8 Steepest descent option not in effect Initial 'Maxtrees' setting = 100 Branches collapsed (creating polytomies) if maximum branch length is zero 'MulTrees' option in effect No topological constraints in effect Trees are unrooted Heuristic search completed Total number of rearrangements tried = 48442 Score of best tree(s) found = 146 Number of trees retained = 2 Time used = 0.01 sec (CPU time = 0.00 sec) paup> DescribeTrees all; Tree description: Unrooted tree(s) rooted using outgroup method Optimality criterion = parsimony Character-status summary: Of 53 total characters: 1 character is of type 'ord' (Wagner) 52 characters are of type 'unord' All characters have equal weight All characters are parsimony-informative Gaps are treated as "missing" Multistate taxa interpreted as uncertainty Character-state optimization: Accelerated transformation (ACCTRAN) Tree number 1 (rooted using user-specified outgroup) Tree length = 146 Consistency index (CI) = 0.3973 Homoplasy index (HI) = 0.6027 Retention index (RI) = 0.6966 Rescaled consistency index (RC) = 0.2767 /----------------------------------------------------------------------------------------------| /-------+-------------------------------------------------------------------------------------31-------32 /-----------------------| /-------------------------------------------------------------30 /---------------| | \------29 /-------| | \------28-------\------33 /------------------------------------------------------------------------------| | /-------\------34 /-------------------------------------------------------------35-------| | /-------------------------------| | /-----------------------------41 /-----------------------\------36 | \------40 /-------| | | /------37-------| | \------39 /-------\------42 \------38-------| /-------| /---------------------------------------------43-------\------44 /----------------------------------------------\------45 /---------------------------------------| | /-------\-----46 /----------------------52-------\------51 /-------| /--------------47-------\------50 /---------------\------49 /-------\------48-------- Lydekkerina Rhineceps Uranocentrodon Benthosuchus Thoosuchus Angusaurus Trematosaurus Wetlugasaurus Vladlenosaurus Odenwaldia Parotosuchus Calmasuchus acri Cyclotosaurus Tatrasuchus Mastodonsaurus Eryosuchus Watsonisuchus Edingerella Xenotosuchus Cherninia Antarctican taxon Paracyclotosaurus St.pronus St.birdi Procycloto Eocyclotosaurus Quasicyclotosaurus Tree number 2 (rooted using user-specified outgroup) Tree length = 146 Consistency index (CI) = 0.3973 Homoplasy index (HI) = 0.6027 Retention index (RI) = 0.6966 Rescaled consistency index (RC) = 0.2767 /----------------------------------------------------------------------------------------------| /-------+-------------------------------------------------------------------------------------31-------32 /-----------------------| /-------------------------------------------------------------30 /---------------| | \------29 /-------| | \------28-------\------33 /------------------------------------------------------------------------------| | /-------\------34 /-------------------------------------------------------------35-------| | /---------------------------------------\------36 /---------------------41 /-------------------------------| | \------40 /-----------------------| | \------39 /---------------\------42 \------38 /-------| \------37-------| /-------| /---------------------------------------------43-------\------44 /----------------------------------------------\------45 /---------------------------------------| | /-------\-----46 /----------------------52-------\------51 /-------| /--------------47-------\------50 /---------------\------49 /-------\------48-------- Input data matrix: 11111111112222222222333333333344444444445555 Taxon 12345678901234567890123456789012345678901234567890123 ------------------------------------------------------------------------Antarctican taxon 0?0??012?0111201110?10?011011110??1111?????????110?11 Angusaurus 11100111110100111011111110001000100110101111100000010 Benthosuchus 11001011010101110011010010001100110111100011000000010 Cherninia 010110020011120100000000111011111111111??????00101?10 Cyclotosaurus 010200020011110012001010111111111111111110???10101000 St.pronus 0101000200111201100000011111111111?1111100?1011001011 Eocyclotosaurus 11021012010112111001111011111110111111???????10001012 Mastodonsaurus 01010012011101011101100011111111111111111111000110010 Paracyclotosaurus 0101100200111201010?100?111111111111111100?1011111?11 Parotosuchus 01001002001112010100000011101111111111110011000000110 Tatrasuchus 0101?002001111000200000011101111111111111111010001100 Thoosuchus 11000111110100110011011110000000100110101111100000010 Trematosaurus 111001111101001110?11111???01001100?10101111100000010 St.birdi 01010002001112011000000111111111111111110011011001111 Rhineceps 00000000000001000000000000000000000001000000000000000 Quasicyclotosaurus 01021012010102101001001011?11110??0111???????10001012 Xenotosuchus 01011002001111011000000011101111?11111110011011100010 Procycloto 1102100200111201100?0???1??111?1111111???????01?01?11 2 Calmasuchus acri 0101000?0011?1?1?10100001?1011111????1110011000000101 Watsonisuchus 01001002001112010010000011101110111111110011011000010 Vladlenosaurus 1100100200011001001001001?10111???????11?11?000000010 Lydekkerina 01100001000101000000000010000100100010100011000000000 Odenwaldia 110010020??1110?0??1????????1???111?11???????00????10 Eryosuchus 01011002001111011100000011101111111111111011000110100 1 Wetlugasaurus 01001002000111010010010011101101011111111011000000010 1 Edingerella 01001002001111100000000010101010010?1101001?011000000 1 1 Uranocentrodon 0000000000000000000000000000010?000001000000000000000 paup> CStatus; Lydekkerina Rhineceps Uranocentrodon Benthosuchus Thoosuchus Angusaurus Trematosaurus Wetlugasaurus Vladlenosaurus Odenwaldia Parotosuchus Calmasuchus acri Tatrasuchus Eryosuchus Cyclotosaurus Mastodonsaurus Watsonisuchus Edingerella Xenotosuchus Cherninia Antarctican taxon Paracyclotosaurus St.pronus St.birdi Procycloto Eocyclotosaurus Quasicyclotosaurus Character-status summary: Current optimality criterion = parsimony No characters are excluded Of 53 total characters: 1 character is of type 'ord' (Wagner) 52 characters are of type 'unord' All characters have equal weight All characters are parsimony-informative paup> Log start; Logging output to file "~/Documents/Manuscripts/ANT Manuscripts/Antarctosuchus MS/Fortuny cladistics/Seb Modified 21 Feb.log".