Folding knotted proteins in a nano-cage - CCP
advertisement

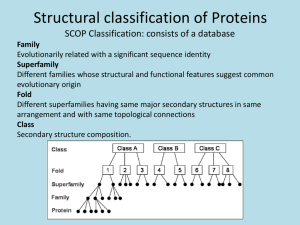
Folding knotted proteins in a nano-cage Joanna I. Sulkowska 1,2 1. Centre of New Technologies, University of Warsaw, Banacha 2c, Warsaw, Poland 2. Faculty of Chemistry, University of Warsaw, Pasteura 1, Warsaw, Poland Thermodynamic properties of middle size knotted proteins are still not known. Recent results have shown that it is possible to determine the free energy landscape of the smallest knotted protein, MJ0366 from Methanocaldococcus jannaschii, with structure based models [1]. On the other hand, recent experimental results have shown that also a bigger protein with much deeper knot, YibK, can tie itself, as also observed in numerical simulations, however chaperonin-assisted folding can significantly speed this process up. In the work [2] we examine the chaperonin-assisted folding and thermodynamics of four knotted proteins by means of molecular dynamics with structures based model. We find that assisted folding significantly increases the probability of knotting for 2efv (which can be fold in the bulk) and allows folding of other small knotted proteins (2rh3 and 4lrv) for which thermodynamics is not accessible in the bulk. Our data show that encapsulation smoothens the free energy landscapes, by omitting kinetic traps and backtracking during knotting mechanism. Moreover, the size of the confinement has a significant impact on the kinetics of folding-unfolding processes, which was also previously reported for proteins with an unknotted topology. Those results suggest that a nano-cage could have a significant influence on knotting probability for middle size proteins as was suggested experimentally for YibK. 1. Noel JK, Onuchic JN, Sulkowska JI (2013), “Knotting a protein in explicit solvent”, The Journal of Physical Chemistry Letters, 4(21), 3570-3573. 2. Niewieczerzał Sz, Sulkowska JI (2014), “Folding knotted proteins in a nano-cage” submitted