Galaxy-History_5-Drug Target Analysis
advertisement

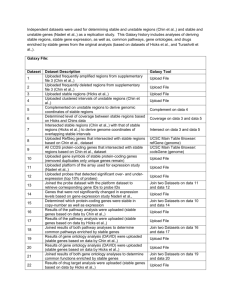
Determined which drugs have targets that are encoded by genes in stable regions (for all breast cancer drugs and comprehensive list of all drugs from DrugBank). Galaxy File: Dataset Dataset Description Uploaded all drugs and drug targets from DrugBank 1 database 2 Uploaded all coding genes associated with stable regions Compared drug targets of drugs with stable genes (which 3 genes are similar between both sets) Joined both drug data and stable gene data indexed by 4 gene symbol, keeping all lines from both queries to retrieve drug information of a stable drug target Joined both drug data and stable gene data indexed by 5 gene symbol, excluding rows that do not join to retrieve drug information of a stable drug target Uploaded all drugs and related diseases from PharmGKB 6 database Uploaded drug file that contained stable drug targets and 7 drug (text in lower case) Joined drug target file with PharmGKB drug relationship file to retrieve more information about the drugs that are 8 associated with stable drug targets (i.e. Pathways and disease information) Removed duplicates based on stable drug target, the drug 9 target (gene name) and the drug. The result was 2,984 unique drugs targeting unique stable genes Counted the number of occurrences of each drug - i.e. 10 How many stable drug targets for each drug Joined the count of all drugs with data of drug targets indexed by drug name – result: all stable drug targets with 11 associated drug that has a count based on how many stable drug targets it is associated with Uploaded all stable genomic regions (genomic 12 coordinates) 13 14 15 16 17 18 19 20 21 22 Uploaded all known UCSC genes Determined which genes were 100% covered by stable regions Filtered based on the coverage amount (i.e. Genes only that covered 100% by stable regions) Uploaded kgXref data from UCSC Table Browser to retrieve gene names/symbols of each known UCSC gene Uploaded data from DrugBank database which contained a list of all drugs and drug targets Uploaded the list of genes that were found to be stable in copy number as well as expression Joined (including rows that do not join) stable genes with drug targets from drug bank to determine which targets are encoded by stable genes Joined (excluding rows that do not join) stable genes with drug targets from drug bank to determine which targets are encoded by stable genes Counted the number of stable drug targets for each drug Counted the number of all drug targets for each drug Galaxy Tool Upload File Upload File Compare two Queries on data 1 and data 2 Join two Queries on data 1 and data 2 Join two Queries on data 1 and data 2 Upload File Upload File Join two Queries on data 6 and data 7 Upload File Count on data 9 Join two Queries on data 9 and data 10 Upload File UCSC Main Table Browser: knownGene (genome) Coverage on data 12 and data 13 Filter on data 14 UCSC Main Table Browser: kgXref (genome) Upload File Upload File Join two Queries on data 17 and data 18 Join two Queries on data 17 and data 18 Count on data 20 Count on data 19 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 Joined counts of drug targets and stable drug targets indexed by drug name to determine of how many drug targets per drug and how many are stable Uploaded specifically breast cancer drugs and targets that were joined with stable genes to determine which drug targets are stable for each breast cancer drug Uploaded all breast cancer drug targets and associated pathways (KEGG pathway analysis by Webgestalt) Joined pathway information with breast cancer drugs that have stable drug targets Joined list of stable genes with kgXref data to determine gene symbols of each stable gene Formatted list of stable genes (list of gene symbols with associated UCSC Id) Joined UCSC Ids of stable genes with genomic coordinates of all genes to determine genomic coordinates of stable genes Used the count tool on list of stable genes (coordinates) to determine how many stable genes occurred for each autosome Uploaded all stable genes (gene names) Uploaded breast cancer drugs with associated drug targets Joined drug data with stable genes to determine which drugs have stable drug targets Uploaded formatted list of breast cancer drugs with stable drug targets Uploaded drug information from DrugBank which included drugs and drug targets Uploaded all stable protein-coding genes (those genes encoded by stable regions) Joined drug information with stable genes to determine all drugs (not solely those to treat breast cancer) that have targets encoded by stable regions Join two Queries on data 21 and data 22 Upload File Upload File Join two Queries on data 24 and data 25 Join two Queries on data 16 and data 18 Upload File Join two Queries on data 13 and data 28 Count on data 29 Upload File Upload File Join two Queries on data 31 and data 32 Upload File Upload File Upload File Join two Queries on data 35 and 36