Supplementary material Table S1. Oligonucleotide primers used in
advertisement
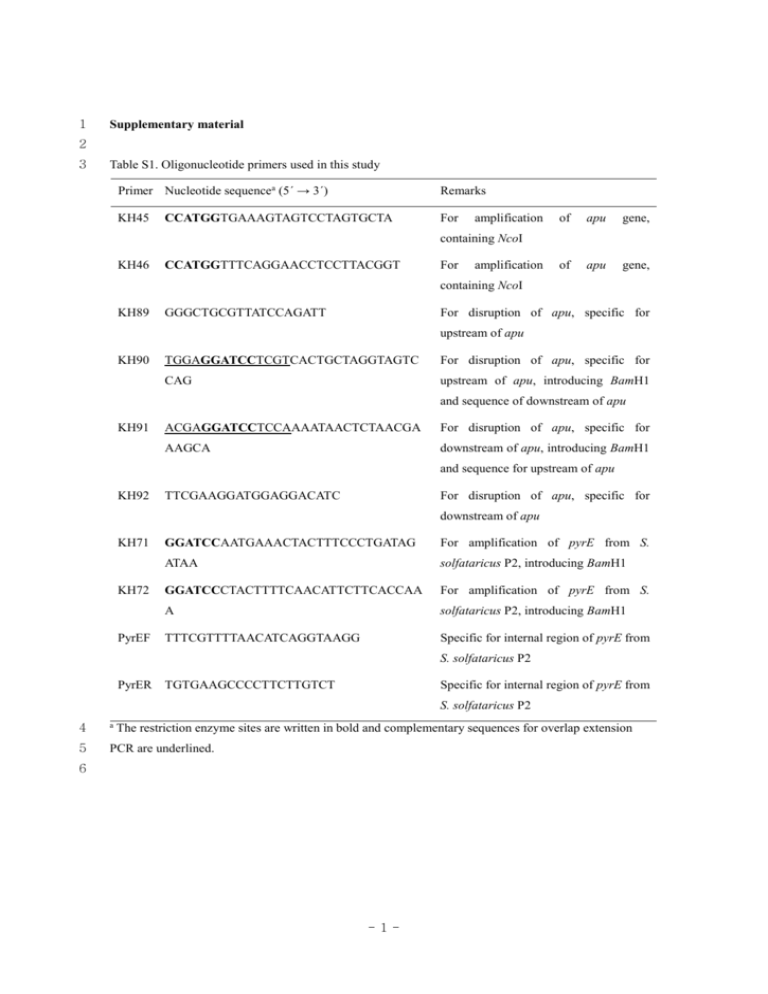
1 Supplementary material 2 3 Table S1. Oligonucleotide primers used in this study Primer Nucleotide sequencea (5´ → 3´) Remarks KH45 For CCATGGTGAAAGTAGTCCTAGTGCTA amplification of apu gene, of apu gene, containing NcoI KH46 CCATGGTTTCAGGAACCTCCTTACGGT For amplification containing NcoI KH89 GGGCTGCGTTATCCAGATT For disruption of apu, specific for upstream of apu KH90 TGGAGGATCCTCGTCACTGCTAGGTAGTC For disruption of apu, specific for CAG upstream of apu, introducing BamH1 and sequence of downstream of apu KH91 ACGAGGATCCTCCAAAATAACTCTAACGA For disruption of apu, specific for AAGCA downstream of apu, introducing BamH1 and sequence for upstream of apu KH92 TTCGAAGGATGGAGGACATC For disruption of apu, specific for downstream of apu KH71 KH72 PyrEF GGATCCAATGAAACTACTTTCCCTGATAG For amplification of pyrE from S. ATAA solfataricus P2, introducing BamH1 GGATCCCTACTTTTCAACATTCTTCACCAA For amplification of pyrE from S. A solfataricus P2, introducing BamH1 TTTCGTTTTAACATCAGGTAAGG Specific for internal region of pyrE from S. solfataricus P2 PyrER TGTGAAGCCCCTTCTTGTCT Specific for internal region of pyrE from S. solfataricus P2 4 a 5 PCR are underlined. The restriction enzyme sites are written in bold and complementary sequences for overlap extension 6 -1- 7 Table S2. Effects of metal ions and reagents on the activity of Apu Reagents Specific activity (U/mg) Relative activity (%) Metal ions None 0.9 100 2+ 1.0 112 Co2+ 1.3 146 Cu2+ 0.9 102 2+ Ca Fe 1.0 113 2+ Mg 1.2 133 Mn2+ 0.9 103 1.0 109 2-mercaptoethanol 1.3 140 Dithiothreitol 1.3 137 1.0 104 2+ Zn Chemical reagents Dimethylaminopropyl ethylcarbodiimide EDTA 1.0 SDS ND Urea 1.0 111 a 109 8 The enzyme was pre-incubated with the above reagents at room temperature for 1 h. Aliquots were tested 9 for Apu activity by incubating the samples with 0.1% soluble starch in 20 mM sodium citrate buffer (pH 10 3.0) at 95 oC for 4 h. The metal ions and other reagents were added at a final concentration of 1 mM except 11 for SDS (1%). The relative values shown are the percentage of the activity without additives. 12 a ND, not detected. 13 14 -2- 15 Table S3. Relative amylolytic activity of Apu on different substrates Substrates Relative activity (%) Polysaccahrides (0.1%) Starch 77.8 Amylose 46.2 Amylopectin 71.8 Glycogen 52.8 Pullulan 100 β-cyclodextrin 2.8 Oligosaccharides (1 mM) Maltose 7.8 Maltotriose 13.8 Maltotetraose 55.2 Maltopentaose 100 Maltohexaose 94.5 Maltoheptaose 86.2 16 The substrates (1 mM or 0.1%) were digested with recombinant Apu (0.2 U) in 20 mM sodium citrate 17 buffer (pH 3.0) for 12 h at 95 oC. The hydrolyzed products were separated on a silica gel TLC plate and 18 quantified using a densitometer (ImageJ software, ver. 1.46, National Institute of Mental Health, Bethesda, 19 MD). 20 -3-