IO3-water-JCP SI rev
advertisement

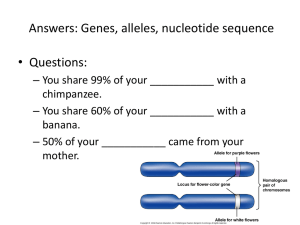
Supplementary Material Solute Anisotropy Effects in Hydrated Anion and Neutral Clusters Hui Wen,1 Gao-Lei Hou,2 Shawn M. Kathmann,1 Marat Valiev, 2,a) Xue-Bin Wang1,a) 1 Chemical & Materials Sciences Division, Pacific Northwest National Laboratory, 902 Battelle Boulevard, P. O. Box 999, Richland, Washington 99352, 2 Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, P. O. Box 999, Richland, Washington 99352 *Correspondence to: xuebin.wang@pnnl.gov; marat.valiev@pnnl.gov Materials and Methods: Photoelectron Spectroscopy A low temperature photoelectron spectrometer coupled to an electrospray ion source and a cryogenic ion-trap was used to obtain photoelectron spectra at 20 K.19 In this work, sodium and potassium iodate salts were dissolved in acetonitrile : water (7:3 v/v) and the above solutions were directly used for spraying. The IO3–(H2O)n cluster anions generated were trapped and cooled for 20-80 ms before being transferred into the extraction zone of a time-of-flight mass spectrometer. These anions were then mass-selected and decelerated before being photodetached with a 157 nm F2 excimer laser. The laser was operated at a 20 Hz repetition rate with the ion beam off at alternating laser shots to enable shot-to-shot background subtraction to be carried S1 out. Photoelectrons were collected at ~100% efficiency with the magnetic bottle and analyzed in a 5.2 m long electron flight tube. The time-of-flight photoelectron spectra were collected and converted to kinetic energy spectra, calibrated using I–, ClO2–, and Cu(CN)2–. The electron binding energy spectra were obtained by subtracting the kinetic energy spectra from the detachment photon energy, and had a resolution (E/E) of ~2% or 20 meV at 1 eV as measured for I– at 355 nm. Electron binding energy was obtained by drawing a straight line along the rising edge of each spectrum and adding instrumental resolution to the crossing point with the binding energy axis. Theoretical Details Calculations were performed using NWChem computational chemistry package.21 We have used 6-31++G** basis set s1 for solvent water molecules. The basis set for iodate consisted of SDB- aug-cc-pVTZ basis for iodine,s2 and cc-pvtz basis set for oxygen atoms.s3 All optimizations were performed at DFT/B3LYP level of theory.s4 Electron binding energies calculated by taking the difference between anion (singlet) and neutral (doublet) species at the optimized anion geometry. For each iodate-water cluster, numbers of different isomers were investigated (up to 30-40 candidates per structure). References S1 J. D. Dill, J. A. Pople, J. Chem. Phys. 62, 2921 (1975). S2 J. M. L. Martin, A. Sundermann, J. Chem. Phys. 114, 3408 (2001). S3 J.T. H. Dunning, J. Chem. Phys. 90, 1007 (1989). S4 A. D. Becke, J. Chem. Phys. 98, 1372 (1993); C. Lee, W. Yang, R. G. Parr, Phys. Rev. B 37, 785 (1988). S2 Table 1S. Electron binding energy (EBE in eV) measured from the maximum of the first spectral feature and calculated from the lowest minimum of each IO3–(H2O)n (n = 0-12) clusters. The threshold detachment energy, TDE, estimated from each spectral threshold is also listed. Number of water molecules, n Experiment Calculation* TDE (EBE) EBE 0 4.70 (4.75) 4.55 1 5.21 (5.32) 5.15 2 5.66 (5.76) 5.62 3 5.85 (5.98) 5.88 4 6.05 (6.27) 6.06 5 6.29 (6.57) 6.30 6 6.53 (6.83) 6.43 7 6.68 (7.02) 6.56 8 6.92 (7.29) 6.68 9 7.12 (7.54) 6.90 10 6.97 (7.38) 6.85 11 7.02 (7.45) 7.01 12 7.12 (7.50) 7.10 *Calculated from the lowest isomer for each cluster shown in Fig.2. The other low lying isomers and their EBEs are given in Figs. 1S-11S. S3 FIG. 1S: Lowest isomers for n=2 cluster S4 FIG. 2S: Lowest isomers for n=3 cluster S5 FIG. 3S: Lowest isomers for n=4 cluster S6 FIG. 4S: Lowest isomers for n=5 cluster S7 FIG. 4S: Lowest isomers for n=5 cluster S8 FIG. 5S: Lowest isomers for n=6 cluster S9 FIG. 5S: Lowest isomers for n=6 cluster S10 FIG. 5S: Lowest isomers for n=6 cluster S11 FIG. 5S: Lowest isomers for n=6 cluster S12 FIG. 6S: Lowest isomers for n=7 cluster S13 FIG. 6S: Lowest isomers for n=7 cluster S14 FIG. 7S: Lowest isomers for n=8 cluster S15 FIG. 7S: Lowest isomers for n=8 cluster S16 FIG. 8S: Lowest isomers for n=9 cluster S17 FIG. 8S: Lowest isomers for n=9 cluster S18 FIG. 9S: Lowest isomers for n=10 cluster S19 FIG. 9S: Lowest isomers for n=10 cluster S20 FIG. 9S: Lowest isomers for n=10 cluster S21 FIG. 10S: Lowest isomers for n=11 cluster S22 FIG. 10S: Lowest isomers for n=11 cluster S23 FIG. 10S: Lowest isomers for n=11 cluster S24 FIG. 11S: Lowest isomers for n=12 cluster S25 FIG. 11S: Lowest isomers for n=12 cluster S26 FIG. 11S: Lowest isomers for n=12 cluster S27