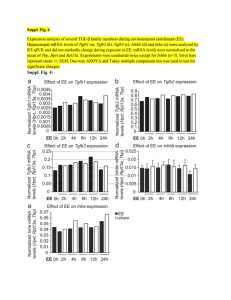
Suppl Fig
advertisement

Suppl Fig. 1 ICSBP expression in different subgroups of AML compared to a common reference RNA; target: AI391632 (100% homology to ICSBP). a) and b) The data are extracted from gene expression data of Bullinger et al., 2010 (http://www.ncbi.nlm.gov/geo, accession number GSE425) at Gene Expression Omnibus (Edgar et al., 2002) and Bullinger et al., 2007 at Gene Expression Omnibus (http://www.ncbi.nlm.gov/geo, accession number GSE8653) using GEOquery (Davis and Meltzer, 2007) on Bioconductor (Gentlemen et al., 2004). The bar plot is generated using the barplot function from the graphics package of R 2.11.1 (R Development Core Team, 2010). c) Boxplot of the log2 ratios of the cytogenetical defined subgroups shown in a); analysed by Mann-Whitney using Wilcoxon rank sum test of R (R Development Core Team, 2010). Suppl Fig. 2 a) Probe mixture: The ICSBP probe (rp11-542M13) is labelled by a green fluorescence dye and the flanking probes (rp11-442O1 ans rp11-158I3) by a red fluorescence dye, resulting in a fusion signal. b) FISH on normal metaphases. Arrows indicate the two fusion signals in 16q24.1. c) Cells of a patient with MDS present two fusion signals. Suppl Fig. 3 SNP data were obtained from Gene Expression Omnibus (Edgar et al., 2002) at http://www.ncbi.nlm.nih.gov/geo. The GSE23452 record containing SNP data of AML patients (Parkin et al. 2010) and the GSE23300 record containing SNP data of MDS patients (Smith et al.) was analyzed. On the analysis of GSE23452, patients 1 and 211 where not included because of the lack of a correlate normal sample. The plots of six patients with loss of the locus 16q24.1 (on the right side) compared to normal tissue (on the left side) are shown. Copy number estimation of the Affymetrix 6.0 SNP array data was done using crlmm, version 1.8.4 (Scharpf et al. 2011) and SNPchip, version 1.14.0 (Scharpf et al. 2007 and 2008) on R, version 2.12.1 (R Development Core Team, 2010) on a 64 bit Linux System containing 8 GB of RAM, according to the copy number vignette of crlmm. Suppl Fig. 4 Shown are representative pyrogrammes of patients P19, P46, P47, and P96 presenting a methylated MSP product. The sequence in the upper part of each Pyrogram represents the sequence under investigation. The sequence below the Pyrogram indicates the sequentially added nucleotides. The gray regions highlight the analyzed C/T sites, with percentage values for the respective cytosine above them . Suppl Fig. 5 a) Methylation-specific PCR demonstrates ICSBP promoter methylation (m) in all investigated cell lines except ME-1, MV4-11, and U937. Except for the cell lines HL-60, Kasumi-1and Me-1, unmethylated (u) ICSBP promoter sequences are also present. 100bp DNA ladder used as standard. b) Western blot analysis using an anti-ICSBP antibody (~50 kDa) and anti-b actin as loading control (42 kDa) shows that ICSBP protein is expressed in MV4-11, U937, and NB4.